Introduction
Breast cancer (BC) is one of the most common malignancies and the most diagnosed neoplasm among women globally (Ref. 1). The 5-year survival rate for BC varies widely depending on the stage of the disease at diagnosis. The estimated 5-year survival for patients with BC diagnosed at Stages I and II ranges from 92% to 100% and decreases drastically to 74% at Stage III and 23% at Stage IV (Ref. Reference Ellison and Saint-Jacques2). The substantial increase in mortality as cancer progresses suggests the importance of early-stage diagnosis and personalised disease management, which has the potential to increase survival rates significantly.
BC diagnostics often involve multiple techniques, such as mammography, magnetic resonance imaging and ultrasound. While these methods are widely applied, they have limitations, such as a high probability of false positives or negatives (Ref. Reference Wu and Chu3). Additionally, the gold standard for BC diagnosis is tumour tissue biopsy; however, this technique is invasive, can cause a risk of infection and can yield inconclusive results due to tumour heterogeneity and potential sample size inadequacy (Ref. Reference Wu and Chu3).
Liquid biopsy has emerged as a credible alternative to traditional diagnostic methods, offering numerous benefits. It involves the collection of a sample of blood followed by the analysis of its components, which include circulating tumour cells (CTCs), cell-free/circulating tumour DNA (cf/ctDNA), exosomes, microRNA (miRNA) and proteins (Refs Reference Cai, Zhang, He, Xia, Dong, Chen, Zhou, Hu, Zhong, Wang, Chen, Xie, Liu and Liu4, Reference Shi, Wartmann, Accuffi, Al-Madhi, Perrakis, Kahlert, Link, Venerito, Keitel-Anselmino, Bruns, Croner, Zhao and Kahlert5, Reference Hsu, Su, Rittenhouse-Olson, Attwood, Mojica, Reid, Dy and Wu6, Reference Alba-Bernal, Godoy-Ortiz, Domínguez-Recio, López-López, Quirós-Ortega, Sánchez-Martín, Roldán-Díaz, Jiménez-Rodríguez, Peralta-Linero, Bellagarza-García, Troyano-Ramos, Garrido-Ruiz, Hierro-Martín, Vicioso, González-Ortiz, Linares-Valencia, Velasco-Suelto, Carbajosa, Garrido-Aranda, Lavado-Valenzuela, Álvarez, Pascual, Comino-Méndez and Alba7). The technique’s clinical benefits include its minimally invasive nature, the ability to perform serial sampling and the ability to generate a systemic picture of (epi)genomic changes in various tumour foci and metastases. This method allows real-time monitoring of tumour progression dynamics and potentially informs on the presence of minimal residual disease (Figure 1) (Ref. Reference Wang, Wang, Lin, Zhu, Huang, Lai, Xi, Huang, Zhang and Zhong8).
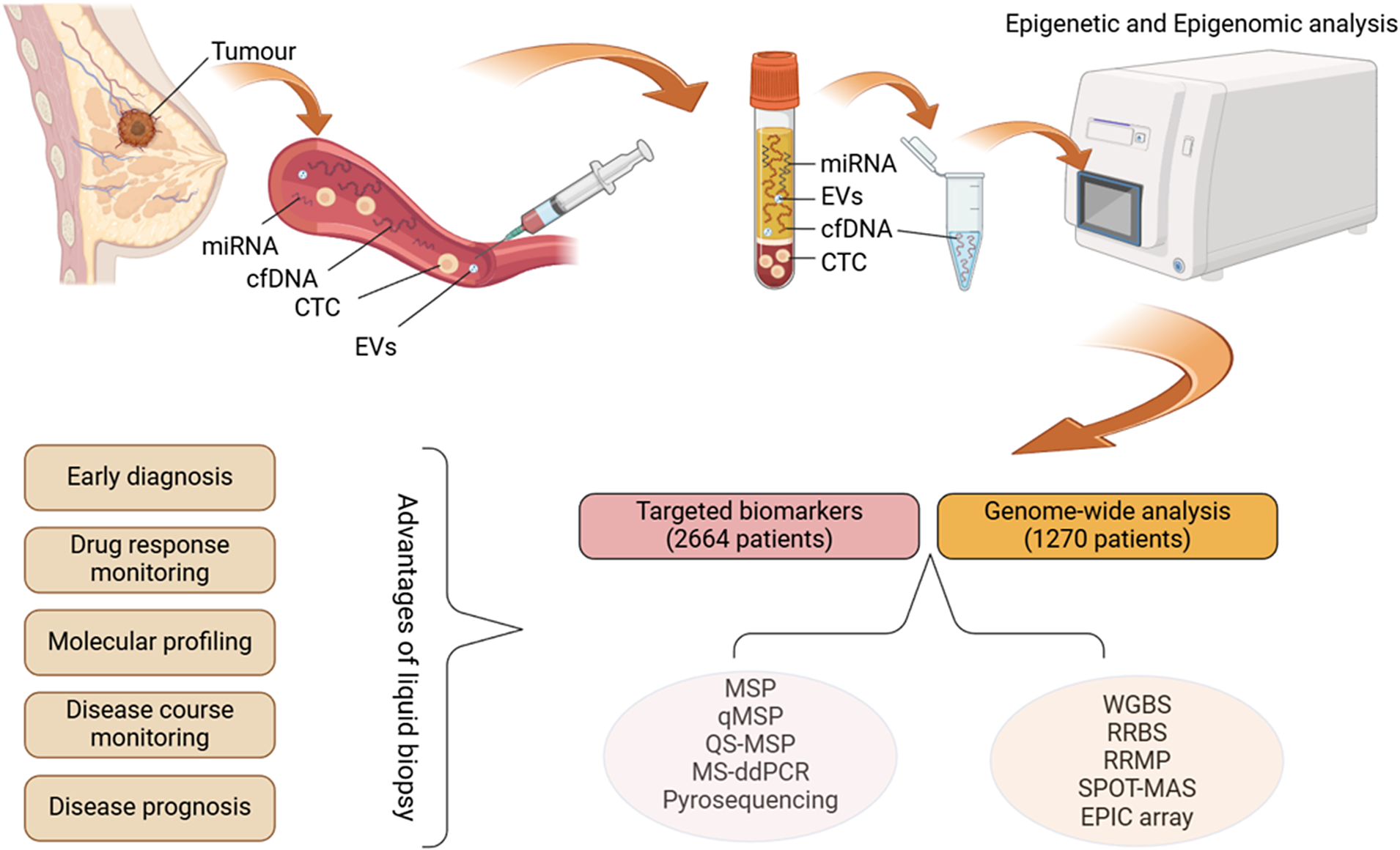
Figure 1. Liquid biopsy workflow and the methods used in reviewed articles (Created with BioRender.com). cfDNA: cell-free DNA; CTC: circulating tumour cell; EV: extracellular vesicles; miRNA: microRNA; MSP: methylation-specific PCR; qMSP: quantitative methylation-specific PCR; OS-MSP: one-step methylation-specific PCR; MS-ddPCR: methylation-specific droplet digital PCR; WGBS: whole-genome bisulphite sequencing; RRBS: reduced representation bisulphite sequencing; RRMP: reduced representative methylome profiling; SPOT-MAS: screening for the presence of tumours by DNA methylation and size.
Most of the research on liquid biopsies has focused on differentiating gene mutational profiles between cancer patients and healthy controls. However, liquid biopsy is also a valuable tool for analysing epigenetic changes. Epigenetic changes include DNA and histone modifications as well as non-coding RNAs (ncRNA). One of the major epigenetic alterations involves DNA methylation – a widespread modification in which a methyl group is added to CpG dinucleotides (Ref. Reference Palanca-Ballester, Rodriguez-Casanova, Torres, Calabuig-Fariñas, Exposito, Serrano, Redin, Valencia, Jantus-Lewintre, Diaz-Lagares, Montuenga, Sandoval and Calvo9). DNA methylation is an essential process in regulating gene transcription and neoplasm formation. Hypermethylation of specific loci, such as tumour suppressors, has been widely observed in cancer tissues, leading to the downregulation of the expression of these genes (Ref. Reference Lakshminarasimhan and Liang10). However, global genomic hypomethylation has also been recognised as a key driver of tumorigenesis, leading to mobile genomic elements and oncogene activation, thus accelerating tumour progression and metastatic lesion formation (Ref. Reference Das and Singal11). These methylation patterns can often be detected through liquid biopsy by analysing cell-free DNA (cfDNA), which shows promise in facilitating early diagnosis and predicting tumour response to therapy (Ref. Reference Palanca-Ballester, Rodriguez-Casanova, Torres, Calabuig-Fariñas, Exposito, Serrano, Redin, Valencia, Jantus-Lewintre, Diaz-Lagares, Montuenga, Sandoval and Calvo9).
Since 1999, when Wong et al. and Esteller et al. discovered cancer-related aberrant DNA methylation in patients’ serum, the importance of DNA methylation research in liquid biopsies has markedly increased, and the list of biomarkers is constantly growing (Refs Reference Wong, Lo, Zhang, Liew, Ng, Wong, Lai, Lau, Hjelm and Johnson12, Reference Esteller, Sanchez-Cespedes, Rosell, Sidransky, Baylin and Herman13). Examples of such cfDNA biomarkers for BC are the ESR1 and SFN genes, whose promoters are hypermethylated in BC and enable credible differentiation of BC patients from healthy controls (Ref. Reference Martínez-Galán, Torres, Del Moral, Muñoz-Gámez, Martín-Oliva, Villalobos, Núñez, Luna Jde, Oliver and Ruiz de Almodóvar14). High PTEN methylation has been associated with the late stages of BC, suggesting that PTEN methylation is a prognostic biomarker (Ref. Reference Swellam, Saad, Sabry, Denewer, Abdel Malak and Abouzid15). Additionally, aberrant methylation of TMEM240 has been correlated with poor response to hormone therapy in BC patients (Ref. Reference Lin, Su, Lin, Thi Anh Thu, Liew, Chen, Tzeng, Liu, Chang, Lee and Hung16). The results of such studies highlight the potential of liquid biopsy for improving BC diagnostics and prognostics and predicting patient response to treatment. Moreover, DNA methylation is a stable and easily detectable change, allowing us to analyse it in tumour genetic material and liquid biopsy (Ref. Reference Chen, Li, Zhou, Yao, Liu, Wu and Su17). On the other hand, epigenetic profiling could provide several benefits to overcome limitations associated with mutation-based liquid biopsy analysis. For example, genetic mutations in cancer can be rare and difficult to predict, whereas DNA methylation changes can be more abundant and more accessible to detect (Ref. Reference Johnston, Ross, Ma, Fung and Locke18).
Similarly to DNA methylation changes, variations in certain ncRNA molecules, such as miRNA, long non-coding RNA and circular RNA, have been shown to have both diagnostic and prognostic potential in BC (Refs Reference Wang, Tan, Hu, Liu, Wu, Zheng, Wang, Luo, Wang, Liu, Lu and Tu19, Reference Mahmoud, Sanad, Elshimy and Hamdy20, Reference Yin, Yan, Fang, Guo, Xiong and Zhang21, Reference Qattan, Al-Tweigeri, Alkhayal, Suleman, Tulbah and Amer22). Biomarkers such as miR-21, LINC00511 and hsa_circ_0001785 were associated with BC progression and metastasis (Refs Reference Wang, Tan, Hu, Liu, Wu, Zheng, Wang, Luo, Wang, Liu, Lu and Tu19, Reference Mahmoud, Sanad, Elshimy and Hamdy20, Reference Yin, Yan, Fang, Guo, Xiong and Zhang21). Moreover, several miRNAs were upregulated in triple-negative breast cancer (TNBC) and correlated with poor survival, thus supporting the importance of ncRNA analysis alongside cfDNA studies (Ref. Reference Qattan, Al-Tweigeri, Alkhayal, Suleman, Tulbah and Amer22).
In liquid biopsies, simple PCR-based methods allow fast and reliable biomarker detection, circumventing the need for time-consuming and expensive genomic analysis of cfDNA. However, based on the universal nature of epigenetic changes, tumour type specificity of such biomarkers can be quite low. Further efforts are needed to identify BC-specific, informative and reliable sets of biomarkers suited for clinical application.
This article reviews the existing research on DNA methylation biomarkers in BC obtained through liquid biopsy by providing specific examples, biomarker categories and potential in BC diagnosis, prognosis and prediction of treatment responses.
Methods
Although interest in BC-specific cfDNA methylation analysis has increased more than twofold since 2016, the search for publications was performed on the PubMed database (https://pubmed.ncbi.nlm.nih.gov/) without time limits, covering the period from inception (1998) to 20 April 2024. The following descriptors were used for database searches: (“breast neoplasms”[MeSH Terms] OR (“breast”[All Fields] AND “neoplasms”[All Fields]) OR “breast neoplasms”[All Fields] OR (“breast”[All Fields] AND “cancer”[All Fields]) OR “breast cancer”[All Fields]) AND (“cell free nucleic acids”[MeSH Terms] OR (“cell free”[All Fields] AND “nucleic”[All Fields] AND “acids”[All Fields]) OR “cell free nucleic acids”[All Fields] OR (“cell”[All Fields] AND “free”[All Fields] AND “dna”[All Fields]) OR “cell free dna”[All Fields]) AND (“dna methylation”[MeSH Terms] OR (“dna”[All Fields] AND “methylation”[All Fields]) OR “dna methylation”[All Fields]). The literature search generated 316 results. Appropriate studies were selected through a two-step publication analysis: screening by title and abstract, and full-text analysis leading to the final 39 publications suitable for review analysis (Figure 2).
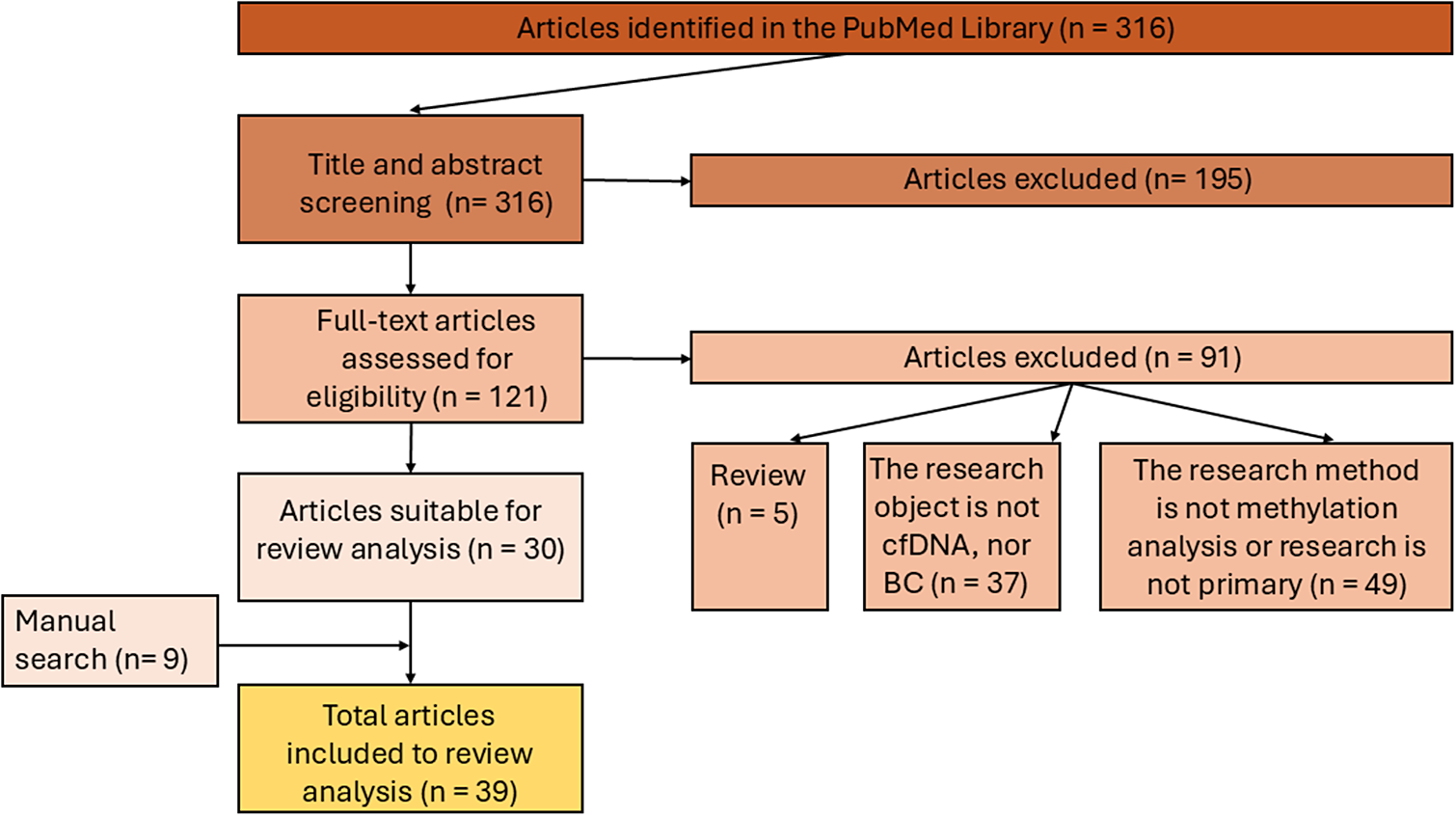
Figure 2. The inclusion/exclusion chart and search method related to the review.
Inclusion criteria
BC cfDNA methylation analysis in liquid biopsies (blood, plasma and serum) and the English language were the primary selection criteria for the papers. The dataset used in this review comprised demographic data from the study group, BC subtypes, analysis techniques, methylated biomarker sensitivity (the proportion of true positive cases in the analysed BC patients’ cohort) and specificity (the proportion of true negative cases in the cohort of non-cancerous samples, and diagnostic or prognostic values) (Supplementary Tables S1 and S2).
Exclusion criteria
Excluded articles were either irrelevant to the topic or study objective; incomplete descriptions of the methods or study groups were also reasons for exclusion. All reviews, letters, case studies, conference material and cohort analyses performed from non-primary studies were excluded from the present analysis.
Considering the selection criteria, 31 articles on target biomarkers and 8 articles analysing the methylome were used for further analysis.
Analysis of the identified biomarkers using the DAVID bioinformatics tool
The Database for Annotation, Visualization, and Integrated Discovery (DAVID; https://davidbioinformatics.nih.gov/) was performed on the analysed biomarker sets (Refs Reference Sherman, Hao, Qiu, Jiao, Baseler and Lane23, Reference Huang, Sherman and Lempicki24). The Gene–Disease Association Database (GAD) was employed to categorise the biomarkers according to their known involvement in human disease categories. The functional annotation analysis associated methylated biomarkers with various biological processes. Furthermore, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed to identify which key signalling pathways the identified biomarkers involved.
The DAVID analysis tool included all biomarkers of Set 1 (51 of targeted analysis) (Refs Reference Cai, Zhang, He, Xia, Dong, Chen, Zhou, Hu, Zhong, Wang, Chen, Xie, Liu and Liu4, Reference Martínez-Galán, Torres, Del Moral, Muñoz-Gámez, Martín-Oliva, Villalobos, Núñez, Luna Jde, Oliver and Ruiz de Almodóvar14-Reference Lin, Su, Lin, Thi Anh Thu, Liew, Chen, Tzeng, Liu, Chang, Lee and Hung16, Reference Ramadan, Hashim, Abouzid and Swellam25-Reference Van der Auwera, Elst, Van Laere, Maes, Huget, van Dam, Van Marck, Vermeulen and Dirix33, Reference Papadopoulou, Davilas, Sotiriou, Georgakopoulos, Georgakopoulou, Koliopanos, Aggelakis, Dardoufas, Agnanti, Karydas and Nasioulas34-Reference Bos, Deger, Sleijfer, Martens and Wilting40, Reference Chimonidou, Tzitzira, Strati, Sotiropoulou, Sfikas, Malamos, Georgoulias and Lianidou41-Reference Panagopoulou, Drosouni, Fanidis, Karaglani, Balgkouranidou, Xenidis, Aidinis and Chatzaki51) and Set 2 (31 of genome-wide analysis) (Refs Reference Widschwendter, Evans, Jones, Ghazali, Reisel, Ryan, Gentry-Maharaj, Zikan, Cibula, Eichner, Alunni-Fabbroni, Koch, Janni, Paprotka, Wittenberger, Menon, Wahl, Rack and Lempiäinen52-Reference Pham, Phan, Jasmine, Tran, Huynh, Vo, Nai, Tran, Truong, Tran, Nguyen, Nguyen, Nguyen, Le, Nguyen, Nguyen, Truong, Do, Phan, Giang, Nguyen and Tran58, Reference Liu, Zhang, Gong, Rugo, Chen, Fu, Che, Tie, Shao, Wan, Kong, Song, Jiang, Xu and Li59), respectively.
Results
Gene-targeted DNA methylation biomarkers for liquid biopsy
Of 31 publications on targeted BC biomarker research, 47% included data from all BC stages, 19% from nonmetastatic (Stages 0–III) BC, and 6% from metastatic Stage IV disease. In 25% of the studies, staging was not specified. More than half of the studies were performed on plasma biosamples (55%), and 45% were performed on serum. The predominant study methods were quantitative methylation-specific PCR and methylation-specific PCR, used in 62.5% and 25% of the studies. DNA extraction in 34% of the studies was performed using a QIAamp DNA Blood Mini Kit (Qiagen), 22% was performed using a QIAamp Circulating Nucleic Acid Kit (Qiagen), and only one study used the standard phenol-chloroform-ethanol method (3%).
Analysis of 31 selected research articles on methylation biomarkers in BC liquid biopsies revealed 51 analysed biomarkers representative of BC. The most studied of all 51 biomarkers were RASSF1 methylation, described in 13 studies, and ESR1 methylation, described in 7 different studies. APC and RARB gene methylation were analysed in six and five studies, respectively, and the methylation of the ATM, FOXA1, MLH1, ITIH5, NBPF1, CDKN2A and PTEN biomarkers was investigated in two studies (Supplementary Table S1).
In these studies, the highest sensitivity and specificity measures for detecting BC were identified at least in one study for SMAD4, PTEN, RARB, APC and DAPK1 gene methylation (Table 1). The sensitivity of these biomarkers was quite high, and the specificity for all of them reached 100% in at least one study.
Table 1. The BC biomarkers’ highest specificity and sensitivity values were reported in individual studies
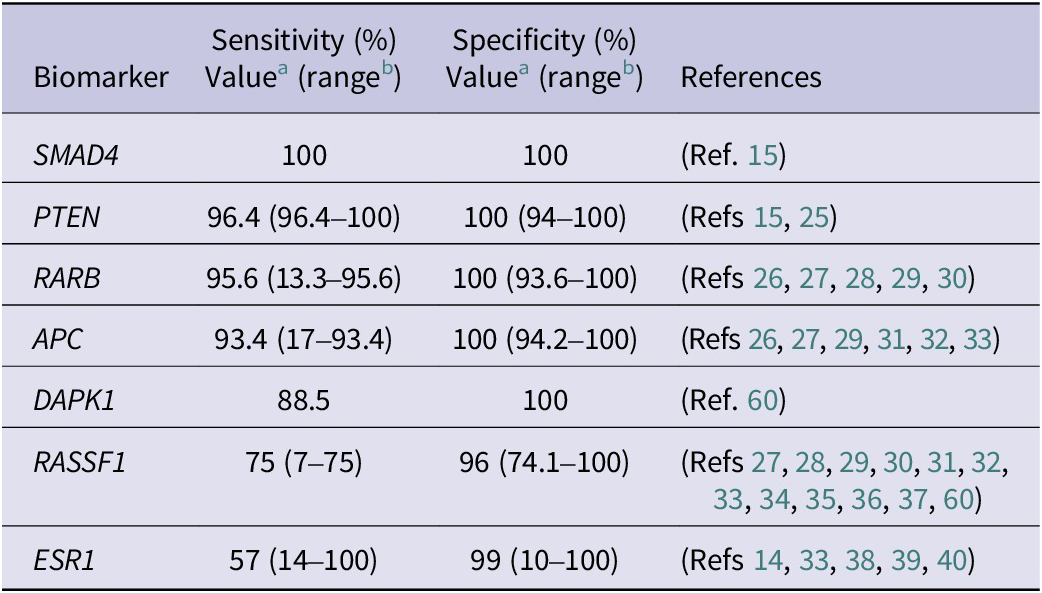
a The highest value of biomarker’s sensitivity and specificity out of all reviewed studies.
b The range of biomarker’s sensitivity and specificity in all reviewed studies.
RASSF1 (Ras Association Domain Family Member 1) methylation is one of the most frequently analysed alterations in BC with relatively high (74%–100%) specificity, albeit with a wide range of sensitivity values (7%–75%) (Table 1). RASSF1 protein is involved in cell cycle control, apoptosis regulation and microtubule stabilisation. It functions as an inhibitor of mitosis that stops the cell cycle at metaphase, which is essential for the correct alignment of chromosomes at the metaphase plate (Ref. Reference Dubois, Bergot, Zalcman and Levallet61). Along with other gene candidates (APC, CCND2, FOXA1, PSAT1 and SCGB3A1), RASSF1 can enhance BC detection accuracy by up to 94% (Ref. Reference Salta, P Nunes, Fontes-Sousa, Lopes, Freitas, Caldas, Antunes, Castro, Antunes, Palma de Sousa, Henrique and Jerónimo32). RASSF1 methylation, which is specific to BC, has an additional advantage as a tool for monitoring the efficacy of neoadjuvant therapy (Ref. Reference Avraham, Uhlmann, Shperber, Birnbaum, Sandbank, Sella, Sukumar and Evron62). In addition, it is predictive of poor overall survival (OS) and disease-free survival (Refs Reference Göbel, Auer, Gaugg, Schneitter, Lesche, Müller-Holzner, Marth and Daxenbichler63, Reference Müller, Widschwendter, Fiegl, Ivarsson, Goebel, Perkmann, Marth and Widschwendter64). The hypermethylation of RASSF1 was found to be associated with hormone receptor-positive (HR-positive) status, as the methylation of RASSF1 is known to be associated with hormone regulation processes (Refs Reference Salta, P Nunes, Fontes-Sousa, Lopes, Freitas, Caldas, Antunes, Castro, Antunes, Palma de Sousa, Henrique and Jerónimo32, Reference Van der Auwera, Elst, Van Laere, Maes, Huget, van Dam, Van Marck, Vermeulen and Dirix33). Moreover, node-positive BC patients exhibit greater RASSF1 methylation levels than node-negative patients; thus, RASSF1 methylation is a biomarker associated with disease progression (Ref. Reference Nunes, Moreira-Barbosa, Salta, Palma de Sousa, Pousa, Oliveira, Soares, Rego, Dias, Rodrigues, Antunes, Henrique and Jerónimo31).
Oestrogen receptor 1 gene (ESR1) methylation is one of the most analysed biomarkers in BC liquid biopsy. The sensitivity of the biomarker varied between 14% and 100%, and the specificity was found to be 10%–100% (Supplementary Table S1). The most promising diagnostic result was detected by analysing the methylation rate of the ESR1 (promoter ER3), which showed 57.5% sensitivity and 99% specificity (Ref. Reference Hagrass, Pasha and Ali39). Bos et al. revealed the opposite result, with 100% sensitivity and only 10% specificity for the ESR1 methylation rate (Ref. Reference Bos, Deger, Sleijfer, Martens and Wilting40).
ESR1 encodes an oestrogen receptor (ER) and ligand-dependent transcription factor that forms homo or heterodimers with ESR2 and has many functions both in reproductive and nonreproductive tissues (Refs Reference Hagrass, Pasha and Ali39, Reference Bos, Deger, Sleijfer, Martens and Wilting40). The ESR1 gene contains multiple promoters and eight exons, leading to transcription product diversity (Ref. Reference Kos, Reid, Denger and Gannon65). While hypermethylation of ESR1 is associated with BC progression (Refs Reference Gerratana, Basile, Franzoni, Allegri, Viotto, Corvaja, Bortot, Bertoli, Buriolla, Targato, Da Ros, Russo, Bonotto, Belletti, Baldassarre, Damante and Puglisi38, Reference Hagrass, Pasha and Ali39), it is not easy to unify ESR1 gene methylation results, as different studies analyse different promoter areas using different methylation analysis methods.
Although increased ER expression is found in approximately 70% of all BC cases, and these patients commonly receive endocrine therapy, it has been shown that some patients are resistant to treatment. Methylation of ESR1 is associated with BC transition from ER+ to ER−, leading to anti-oestrogen treatment resistance and disease progression (Ref. Reference Martínez-Galán, Torres-Torres, Núñez, López-Peñalver, Del Moral, Ruiz De Almodóvar, Menjón, Concha, Chamorro, Ríos and Delgado66).
Although BRCA1 (BRCA1 DNA repair associated) inactivation in BC is usually associated with mutations, methylation of the gene promoter is associated with transcriptional inactivation of BRCA1 and is a second hit for mutation carriers (Ref. Reference Rice, Ozcelik, Maxeiner, Andrulis and Futscher67). BRCA1 methylation is found in 10%–15% of all sporadic BC patients, resulting in complete gene silencing and loss of function (Ref. Reference Rice, Ozcelik, Maxeiner, Andrulis and Futscher67). Loss of BRCA1 in BC cells influences the transformation of luminal progenitor cells to a basal-like BC phenotype (Ref. Reference Lim, Vaillant, Wu, Forrest, Pal, Hart, Asselin-Labat, Gyorki, Ward, Partanen, Feleppa, Huschtscha, Thorne, Fox, Yan, French, Brown, Smyth, Visvader and Lindeman68). BRCA1 is essential for cellular regulation, including apoptosis and genome stability maintenance via DNA repair. BC cells without functional BRCA1 gene (mutated or epigenetically silenced) lose the ability to repair DNA double-strand breaks via a homologous repair pathway. Therefore, double-strand break-inducing therapies, such as platinum-based chemotherapy, result in hypersensitivity to treatment (Ref. Reference Zhu, Shan, Wang, Wang, Wang, Shen, Liu, Wang, Yuan, Ying and Yang69). Studies in ovarian cancer and BC cell lines revealed a similar mechanism of methylated BRCA1 association with poly (ADP-ribose) polymerase inhibitor, where BRCA1 hypermethylation results in an enhanced response to treatment (Refs Reference Kondrashova, Topp, Nesic, Lieschke, Ho, Harrell, Zapparoli, Hadley, Holian, Boehm, Heong, Sanij, Pearson, Krais, Johnson, McNally, Ananda, Alsop, Hutt, Kaufmann, Lin, Harding, Traficante, deFazio, McNeish, Bowtell, Swisher, Dobrovic, Wakefield and Scott70, Reference Villman, Blomqvist, Larsson and Nygren71, Reference Kawachi, amashita, Okochi-Takada, Hirakawa, Tsuda, Shimomura, Kojima, Yonemori, Fujiwara, Kinoshita, Ushijima and Tamura72).
The SMAD4 (SMAD family member 4) biomarker analysed in liquid biopsy showed the highest accuracy, with 100% sensitivity and specificity for BC (Ref. Reference Swellam, Saad, Sabry, Denewer, Abdel Malak and Abouzid15). This biomarker also had prognostic value, while SMAD4 methylation was significantly associated with cancer progression (tumour stage, higher grade and lymph node involvement) (Ref. Reference Swellam, Saad, Sabry, Denewer, Abdel Malak and Abouzid15). It also exhibited an association with HR-positive BC subtypes (Ref. Reference Swellam, Saad, Sabry, Denewer, Abdel Malak and Abouzid15). Moreover, SMAD4 cfDNA methylation correlated with and was superior to clinically used carcinoembryonic antigen (CEA) and cancer antigen 15.3 (CA15.3) biomarkers (Refs Reference Swellam, Saad, Sabry, Denewer, Abdel Malak and Abouzid15, Reference Ramadan, Hashim, Abouzid and Swellam25). While methylated SMAD4 may serve as a biomarker with high sensitivity and specificity for early BC detection, along with methylated PTEN biomarkers, both act as tumour invasiveness and distant metastasis-associated factors (Ref. Reference Swellam, Saad, Sabry, Denewer, Abdel Malak and Abouzid15).
The family of SMAD proteins is responsible for transducing signals within the cell and is involved in numerous signalling pathways. Although SMAD4 loss alone is not linked to carcinogenesis, it interferes with the transforming growth factor beta (TGF-β) and bone morphogenic protein pathways, which can lead to transcriptional activation or inhibition of targeted genes. Additionally, TGF-β/SMAD4 is involved in the DNA damage response and repair by controlling the transcriptional activity of essential genes involved in these processes (Ref. Reference Zhao, Mishra and Deng73). Li et al. reported that SMAD4 is associated with apoptosis in the early stages of ERα-positive BC; therefore, the loss of SMAD4 can induce uncontrolled cell growth due to alterations in cell cycle arrest and apoptosis (Ref. Reference Li, Wu, Oelschlager, Wan, Stockard, Grizzle, Wang, Chen, Sun and Cao74).
Phosphatase and tensin homolog (PTEN) methylation in BC liquid biopsy samples showed 96.4% sensitivity and 100% specificity and was a more potent diagnostic tool than CEA and CA15.3. Furthermore, methylated PTEN correlated with OS (Ref. Reference Ramadan, Hashim, Abouzid and Swellam25). PTEN is a tumour suppressor gene involved in translation, cell cycle and apoptotic processes. PTEN suppresses apoptosis and increases cell survival by negatively regulating the AKT kinase pathway (Ref. Reference Lu, Cheng and Teng75). Moreover, PTEN is involved in DNA repair processes and is essential for BC signalling pathways. The downregulation of PTEN leads to the development of malignant mammary stem/progenitor cells through increased signalling within the AKT/GSK-3β/Wnt/β-catenin pathway; moreover, the loss of PTEN results in resistance to trastuzumab therapy and poor OS (Ref. Reference Esteva, Guo, Zhang, Santa-Maria, Stone, Lanchbury, Sahin, Hortobagyi and Yu76).
RARB (Retinoic Acid Receptor Beta) gene methylation is one of the most frequently analysed epigenetic biomarkers in various cancers (Refs Reference Swellam, Abdelmaksoud, Sayed Mahmoud, Ramadan, Abdel-Moneem and Hefny26, Reference Saeki, Abe, Kono, Nakazato, Ishihara and Abe27, Reference Yamamoto, Nakayama, Kajita, Miyake, Iwamoto, Kim, Sakai, Ishihara, Tamaki and Noguchi28, Reference Hoque, Feng, Toure, Dem, Critchlow, Hawes, Wood, Jeronimo, Rosenbaum, Stern, Yu, Trink, Kiviat and Sidransky29). The RARB is a nuclear receptor and a member of the Retinoic Acid Receptor (RAR) class (Ref. Reference Geoffroy, Esnault and de Thé77). It is a transcription initiator activated by a ligand – a physiologically active form of vitamin A (retinoic acid). The primary function of RARB is to control epithelial cell proliferation and haematopoiesis. RARB is essential for signal transduction pathways, cell division and differentiation processes (Ref. Reference Geoffroy, Esnault and de Thé77).
In all studies analysing BC liquid biopsy samples, RARB methylation was defined as a diagnostic biomarker with the highest sensitivity (95.6%) and 100% specificity (range 12%–95.6% and 94%–100%, respectively) for detecting BC (Table 1). Swellam et al. defined RARB gene methylation as a more robust diagnostic tool than the traditional tumour markers CEA and CA15.3, which are helpful not only for early BC detection but also for early clinical stage, low grade and TNBC definition (90% sensitivity and 100% specificity) (Ref. Reference Swellam, Abdelmaksoud, Sayed Mahmoud, Ramadan, Abdel-Moneem and Hefny26). Kim et al., analysing a set of biomarkers (SCGB3A1, RARB, RASSF1 and TWIST1), reported RARB gene methylation in BC patient serum with a sensitivity of 86.6% and a specificity of 93.6%. Adding to this analysis, a second gene, RASSF1, improved the sensitivity to 94.1% and achieved an area under the curve (AUC) of 0.979, although the specificity was lower (88.8%). Nevertheless, the author proposed that these two gene panels are suitable for early and metastatic BC diagnosis (Ref. Reference Kim, Shin, Kweon, Park, Yoon, Lee, Choi, Fackler and Sukumar30).
APC (APC Regulator of WNT Signalling Pathway) is another frequently analysed methylation biomarker specific to colon cancer that is often hypermethylated in BC specimens with high specificity (94.2%–100%) and various ranges of sensitivity (17%–93.45%) (Table 1). A review of studies analysing APC hypermethylation highlighted APC as a prognostic biomarker associated with advanced tumour stage (Ref. Reference Hoque, Feng, Toure, Dem, Critchlow, Hawes, Wood, Jeronimo, Rosenbaum, Stern, Yu, Trink, Kiviat and Sidransky29), disease progression (Ref. Reference Van der Auwera, Elst, Van Laere, Maes, Huget, van Dam, Van Marck, Vermeulen and Dirix33) and metastasis (Ref. Reference Nunes, Moreira-Barbosa, Salta, Palma de Sousa, Pousa, Oliveira, Soares, Rego, Dias, Rodrigues, Antunes, Henrique and Jerónimo31), as this gene is a crucial cell adhesion-regulating factor (Ref. Reference Saelee and Pongtheerat78). While APC acts as a component of the Wnt/β-catenin signalling pathway, the methylation of APC dysregulates the signalling pathway and increases resistance to chemotherapeutic agents (Ref. Reference Romero-Garcia, Prado-Garcia and Carlos-Reyes79). Another APC study demonstrated that loss of APC function via methylation or mutation acts as an accelerant for ABCB1 gene expression gain, and as a result, cells became resistant to doxorubicin (Refs Reference VanKlompenberg, Leyden, Arnason, Zhang, Stefanski and Prosperi80, Reference Stefanski, Keffler, McClintock, Milac and Prosperi81).
The DAPK1 (Death-associated protein kinase 1) gene encodes calcium- and calmodulin-dependent serine/threonine kinase involved in cell cycle regulation, autophagy, apoptosis, oxidative stress and metastatic processes. DAPK1 is essential for regulating AKT kinase, which is involved in many response pathways that induce metastasis: cell spreading, activation of proliferation, inhibition of apoptosis, regulation of p53 and angiogenesis (Ref. Reference Elbadawy, Usui, Yamawaki and Sasaki82). A high sensitivity (88%) and 100% specificity of DAPK1 gene hypermethylation were shown in one BC cfDNA methylation study (Ref. Reference Van der Auwera, Elst, Van Laere, Maes, Huget, van Dam, Van Marck, Vermeulen and Dirix33). DAPK1 and RARB gene methylation were defined as diagnostic biomarkers, but both showed significant associations with menopausal status (Refs Reference Hoque, Feng, Toure, Dem, Critchlow, Hawes, Wood, Jeronimo, Rosenbaum, Stern, Yu, Trink, Kiviat and Sidransky29, Reference Ahmed, Pusch, Hamed, Rashad, Idris, El-Fadle and Blin60).
DNA methylome profiling in liquid biopsy
In selected publications on liquid biopsy, seven methylome analyses were performed using next-generation sequencing (NGS)-based methods, and one used the EPIC-array method. Three of the eight studies used whole-genome bisulphite sequencing (WGBS) to analyse the whole BC genome. These studies revealed 60,035 differentially methylated regions (DMRs) (Supplementary Table S2). Two studies separated DMRs into hyper and hypomethylated regions, with hypomethylated DMRs comprising the central part of the analysed regions (89% vs. 11%) (Supplementary Table S2). Genome-wide hypomethylation is known to be associated with metastatic BC. In contrast, the early stages of the disease show a hypermethylation profile, indicating a change in the methylation pattern during BC progression (Refs Reference Man, Li, Wang, Zhang, Zhang and Li83, Reference Kar, Sengupta, Deb, Shilpi, Parbin, Rath, Pradhan, Rakshit and Patra84). Genome-wide methylation research revealed 31 BC biomarkers (Supplementary Table S2).
Widschwendter et al. used a reduced representation bisulphite sequencing method to perform ultradeep bisulphite sequencing for primary and metastatic BC to monitor DNA methylation change before and after chemotherapy (Ref. Reference Widschwendter, Evans, Jones, Ghazali, Reisel, Ryan, Gentry-Maharaj, Zikan, Cibula, Eichner, Alunni-Fabbroni, Koch, Janni, Paprotka, Wittenberger, Menon, Wahl, Rack and Lempiäinen52). Ten methylated CpG regions showed the best results and were subsequently optimised to five areas, which covered the DNA methylation marker EFC#93. The hypermethylation of EFC#93 in BC patient serum was a vital marker for both poor relapse-free survival and OS (hazard ratio 5.973). More than 70% of patients who were EFC#93 and CTC-positive relapsed within 5 years, indicating the prognostic potential of this biomarker. Moreover, compared with healthy controls, the methylation-based discrimination of the EFC#93 region in primary and metastatic BC patients showed AUC values of 0.850 and 0.845, respectively. The sensitivity and specificity of this biomarker reached 60.9% and 92%, respectively, when controls were compared to the total group of BC patients. Furthermore, EFC#93 methylation was detected in 43% of women 3–6 months and 25% of them 6–12 months before BC diagnosis with a lethal outcome with 88% specificity and 33.9% sensitivity (fourfold higher than that of nonfatal BC). These results confirm DNA methylation biomarkers’ tremendous prognostic potential and open possibilities for treatment individualisation (Ref. Reference Widschwendter, Evans, Jones, Ghazali, Reisel, Ryan, Gentry-Maharaj, Zikan, Cibula, Eichner, Alunni-Fabbroni, Koch, Janni, Paprotka, Wittenberger, Menon, Wahl, Rack and Lempiäinen52).
Using WGBS data from the two BC cohorts, Liu et al. reported the use of cfMETH (a predictive score based on cfDNA methylation in each sample and computed using a random classifier) combined with diagnostic imaging tools (mammography and ultrasound) to develop diagnostic tests with high sensitivity, specificity and accuracy (95.2%, 78.4%, and 86.8%, respectively, taking the average of both cohorts) for diagnosing BC (Ref. Reference Liu, Zhao, Huang, Xu, Zhou, Zhang, Li, Ming, Wang, Zhao, Li, Dong, Ma, Qian, Chen, Xing, Zhang, Chen, Liu, Pang, Zhou, Wu, Wang, Wang, Wu and Su53). The AUC values of the cfMETH predictive score in the discovery and validation cohorts were 0.89 and 0.81, respectively. When analysing 10 optimal hypo-DMRs distinguishing malignant and benign plasma samples, four genes associated with these DMRs were reported as DNA methylation biomarkers for BC (RYR2, RYR3, GABRB3 and DCDC2C). When comparing WGBS data of the BC genome with those of healthy controls, a hypomethylation pattern at the genome-wide level was found (Ref. Reference Liu, Zhao, Huang, Xu, Zhou, Zhang, Li, Ming, Wang, Zhao, Li, Dong, Ma, Qian, Chen, Xing, Zhang, Chen, Liu, Pang, Zhou, Wu, Wang, Wang, Wu and Su53).
Another WGBS study revealed predominant hypomethylation in BC cfDNA samples (64.5%) and associated 146 genes with differentially methylated CpGs (DMCpGs) and 204 with hypo-DMCpGs (Ref. Reference Luo, Huang, Guo, Guo, Zeng, Li and Liu54). Methylation levels in 13 CpGs were comparable in BC tissue and cfDNA and significantly different between cancerous specimens and noncancerous controls (both in tissues and cfDNA). Thirteen CpGs were described as diagnostic biomarkers and associated with nine genes (Supplementary Table S2). Three of the 13 CpGs were further analysed in an additional cohort. They showed high sensitivity and specificity for diagnosing BC (69.4%–83.7% and 85.7%–88.6%, respectively), indicating that these sites could serve as biomarkers for early-stage BC diagnosis (Ref. Reference Luo, Huang, Guo, Guo, Zeng, Li and Liu54).
Rodriguez-Casanova et al. used the EPIC array method and detected 28,799 DMCpGs in cfDNA from nine metastatic luminal B patients compared to healthy controls (Ref. Reference Rodriguez-Casanova, Costa-Fraga, Castro-Carballeira, González-Conde, Abuin, Bao-Caamano, García-Caballero, Brozos-Vazquez, Rodriguez-López, Cebey, Palacios, Cueva, López-López, Costa and Díaz-Lagares55). As in previous studies, hypomethylated DMCpGs were dominant (92%) and were found in low-density CpG areas (open sea) and outside the promoters. Hypermethylation was predominant in CpG islands and promoters, and 1,467 DMCpGs were generated to differentiate BC from control samples. Thirty-four DMCpGs corresponded to 24 genes associated with the Wnt signalling pathway. Analysis of WNT1 gene hypermethylation revealed a difference in patients with the luminal B subtype of BC versus healthy controls, with an AUC of 0.86, a sensitivity of 78%, and a specificity of 100%. Moreover, researchers are claiming, that WNT1 gene hypermethylation serves not only as a diagnostic BC biomarker but also as a tool for metastasis monitoring (Ref. Reference Rodriguez-Casanova, Costa-Fraga, Castro-Carballeira, González-Conde, Abuin, Bao-Caamano, García-Caballero, Brozos-Vazquez, Rodriguez-López, Cebey, Palacios, Cueva, López-López, Costa and Díaz-Lagares55).
Yang et al. used the reduced representative methylome profiling (RRMP) method. They identified CpGs in the promoter (75.2%) and CpG island areas (81.9%), with the ability to distinguish BC patients from controls, including patients with benign breast lesions and an AUC of 0.85 (Ref. Reference Yang, Zhu, Liu, He, Xu, Zheng, Huang, Wang, Lin, Guo and Chen56). By performing an RRMP analysis on BC lines, the group identified an association between H3K4me3 and hypermethylation, suggesting that this method could be used for histone modification analysis (Ref. Reference Yang, Zhu, Liu, He, Xu, Zheng, Huang, Wang, Lin, Guo and Chen56).
SPOT-MAS (Screening for the Presence Of Tumor by Methylation And Size) refers to a multi-modal liquid biopsy assay that analyzes cell-free DNA in the blood—examining methylation patterns, fragment size profiles, copy-number variations, and end motifs—to detect tumors. Two groups of researchers, including 462 BC patients, performed two SPOT-MAS research analyses (Refs Reference Nguyen, Nguyen, Doan, Pham, Nguyen, Nguyen, Tran, Vo, Phan, Jasmine, Nguyen, Nguyen, Nguyen, Nguyen, Huynh, Tran, Dang, Doan, Tran, Nguyen, Nguyen, Ho, Tran, Pham, Ho, Nguyen, Nguyen, Nguyen, Phu, Phan, Vo, Nai, Tran, Truong, Tran, Le, Tran, Duong, Bach, Kim, Pham, Tran, Le, Pham, Le, Vo, Tran, Nguyen, Van, Nguyen, Tran, Tran, Le, Do, Phan, Nguyen, Nguyen, Cao, Do, Truong, Tang, Giang, Nguyen, Phan and Tran57, Reference Pham, Phan, Jasmine, Tran, Huynh, Vo, Nai, Tran, Truong, Tran, Nguyen, Nguyen, Nguyen, Le, Nguyen, Nguyen, Truong, Do, Phan, Giang, Nguyen and Tran58). In both studies, the cfDNA concentration was higher in cancerous specimens, and short DNA fragments (<150 bp) were associated with BC, indicating that the cfDNA of cancer patients was more fragmented than that of healthy participants. Both studies analysed EMs and found that different 4-mers increased (CA** and GG**) and decreased (CG** and A***) in cancer specimens (Refs Reference Nguyen, Nguyen, Doan, Pham, Nguyen, Nguyen, Tran, Vo, Phan, Jasmine, Nguyen, Nguyen, Nguyen, Nguyen, Huynh, Tran, Dang, Doan, Tran, Nguyen, Nguyen, Ho, Tran, Pham, Ho, Nguyen, Nguyen, Nguyen, Phu, Phan, Vo, Nai, Tran, Truong, Tran, Le, Tran, Duong, Bach, Kim, Pham, Tran, Le, Pham, Le, Vo, Tran, Nguyen, Van, Nguyen, Tran, Tran, Le, Do, Phan, Nguyen, Nguyen, Cao, Do, Truong, Tang, Giang, Nguyen, Phan and Tran57, Reference Pham, Phan, Jasmine, Tran, Huynh, Vo, Nai, Tran, Truong, Tran, Nguyen, Nguyen, Nguyen, Le, Nguyen, Nguyen, Truong, Do, Phan, Giang, Nguyen and Tran58).
Among all the variables included in the SPOT-MAS assay, EM and genome-wide methylation had the best results for cancer detection, with sensitivities of 58.3% and 49.3%, respectively, and specificities >95% in two BC cohorts (Ref. Reference Nguyen, Nguyen, Doan, Pham, Nguyen, Nguyen, Tran, Vo, Phan, Jasmine, Nguyen, Nguyen, Nguyen, Nguyen, Huynh, Tran, Dang, Doan, Tran, Nguyen, Nguyen, Ho, Tran, Pham, Ho, Nguyen, Nguyen, Nguyen, Phu, Phan, Vo, Nai, Tran, Truong, Tran, Le, Tran, Duong, Bach, Kim, Pham, Tran, Le, Pham, Le, Vo, Tran, Nguyen, Van, Nguyen, Tran, Tran, Le, Do, Phan, Nguyen, Nguyen, Cao, Do, Truong, Tang, Giang, Nguyen, Phan and Tran57). The detection rate of BC was the lowest among the five studied cancer types, which could be explained by the low level of cfDNA shedding and molecular subtype heterogeneity (Ref. Reference Liu, Oxnard, Klein, Swanton and Seiden85). By measuring the importance scores of different cfDNA features, the best results for all five cancer types were obtained for BC, with the highest values of 0.87 and 0.78 in the two cohorts, respectively (Ref. Reference Nguyen, Nguyen, Doan, Pham, Nguyen, Nguyen, Tran, Vo, Phan, Jasmine, Nguyen, Nguyen, Nguyen, Nguyen, Huynh, Tran, Dang, Doan, Tran, Nguyen, Nguyen, Ho, Tran, Pham, Ho, Nguyen, Nguyen, Nguyen, Phu, Phan, Vo, Nai, Tran, Truong, Tran, Le, Tran, Duong, Bach, Kim, Pham, Tran, Le, Pham, Le, Vo, Tran, Nguyen, Van, Nguyen, Tran, Tran, Le, Do, Phan, Nguyen, Nguyen, Cao, Do, Truong, Tang, Giang, Nguyen, Phan and Tran57).
Pham et al. performed SPOT-MAS analysis on 239 nonmetastatic BC cfDNA specimens (Ref. Reference Pham, Phan, Jasmine, Tran, Huynh, Vo, Nai, Tran, Truong, Tran, Nguyen, Nguyen, Nguyen, Le, Nguyen, Nguyen, Truong, Do, Phan, Giang, Nguyen and Tran58). Interestingly, as Liu J’s results (Ref. Reference Liu, Zhao, Huang, Xu, Zhou, Zhang, Li, Ming, Wang, Zhao, Li, Dong, Ma, Qian, Chen, Xing, Zhang, Chen, Liu, Pang, Zhou, Wu, Wang, Wang, Wu and Su53) showed increased methylation instability in the TNBC and HER2 BC subtypes, Pham’s work revealed a similar tendency, with the TNBC and luminal B-HER2 subtypes exhibiting a significant proportion of hypermethylation (Ref. Reference Pham, Phan, Jasmine, Tran, Huynh, Vo, Nai, Tran, Truong, Tran, Nguyen, Nguyen, Nguyen, Le, Nguyen, Nguyen, Truong, Do, Phan, Giang, Nguyen and Tran58). Moreover, four regions of DMR covering the SOX17, RASSF1 and OTX2 genes with significant hypermethylation rates distinguished BC patients from healthy individuals (Ref. Reference Pham, Phan, Jasmine, Tran, Huynh, Vo, Nai, Tran, Truong, Tran, Nguyen, Nguyen, Nguyen, Le, Nguyen, Nguyen, Truong, Do, Phan, Giang, Nguyen and Tran58).
Using WGBS, Liu et al. analysed samples from 16 patients with hormone receptor-positive BC (Ref. Reference Liu, Zhang, Gong, Rugo, Chen, Fu, Che, Tie, Shao, Wan, Kong, Song, Jiang, Xu and Li59). The group’s main goal was to analyse ctDNA methylation in patients before and after disease relapse while using exemestane (EXE) therapy. The group identified 79 differential methylation density (MD) regions, which covered 175 genes, and 70 differential methylation ratios (MRs), which covered 223 genes, associated with resistance to EXE. Moreover, the MD and MR regions included seven DMRs in common, which overlapped with various genes participating in the anti-tumour immune response (HLA) or apoptosis and cell cycle regulation (TRIM42) (Supplementary Table S2). Interestingly, according to the MD and MR data, regions covering the genes SUCLG2-DT1, CLSTN2, CLSTN2-AS1, TRIM42 and ANO4 were associated with prognosis for progression-free survival (PFS) after EXE treatment. When predicting EXE resistance, changes in MD and MR in the region covering HLA class II α chain paralogous family genes (HLA-DRA, HLA-DRB1, HLA-DRB5 and HLA-DRB6) were detected, and a greater methylation rate of this region was associated with shorter PFS (Ref. Reference Liu, Zhang, Gong, Rugo, Chen, Fu, Che, Tie, Shao, Wan, Kong, Song, Jiang, Xu and Li59).
The function and importance of selected biomarkers
The DAVID bioinformatics tool (Refs Reference Sherman, Hao, Qiu, Jiao, Baseler and Lane23, Reference Huang, Sherman and Lempicki24) included all 51 targeted hypermethylation biomarkers (Set 1) for further analysis. Concerning the association of Set 1 with human disease, 42 of the 51 biomarkers (82.4%) were associated by GAD disease classification to oncological (54.9%), metabolic (49.0%), neurological (41.2%), cardiovascular system (41.2%) or immune (31.4%) disorders. The same analysis used genome-wide methylation data of 31 BC characteristic biomarkers (Set 2). Analysis revealed that 74.2% of Set 2 biomarkers were associated with metabolic (54.8%), neurological (48.4%), cardiovascular (38.7%), immune (35.5%) and haematological (29.0%) diseases.
In further analysis, Set 1 biomarkers were associated with cellular processes (74.5%). The biomarkers were involved in the cell cycle (17.6%), lipid metabolism (11.8%) and apoptosis (9.8%), while most biomarkers were essential for transcription-associated processes (27.5%) (Figure 3A). Moreover, seven biomarkers identified in the Set 2 analysis were related to two cellular transport-associated processes (Figure 3B).
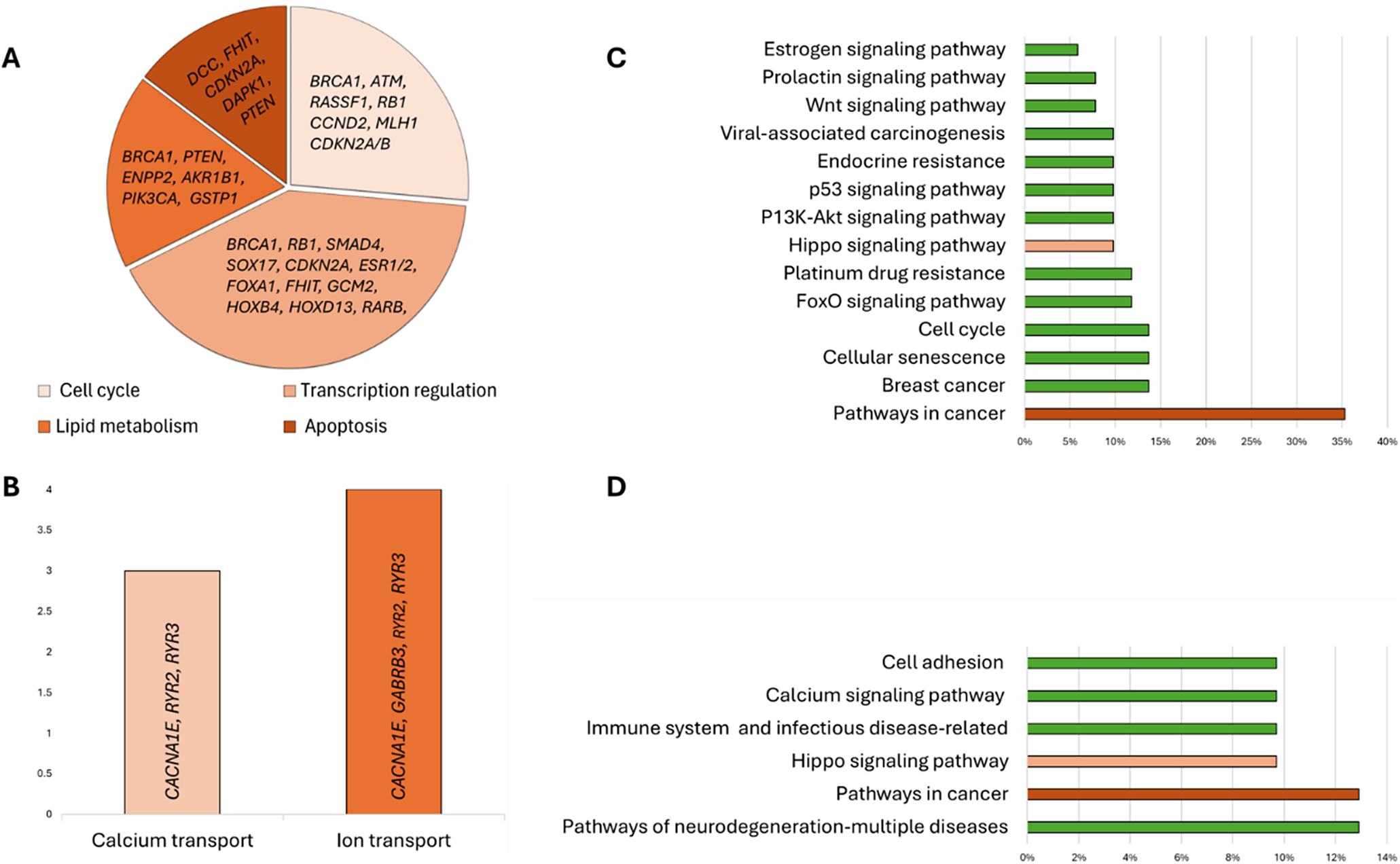
Figure 3. Distribution of biomarkers by their associations with the main cellular processes (A and B) and signalling pathways (C and D). Set 1 (A and C) and Set 2 (B and D) biomarkers’ analysis.
The participation of the targeted biomarkers in various signalling pathways was analysed using KEGG pathway analysis, which revealed that 68.6% of the analysed Set 1 genes participated in different signalling pathways, with most of the genes (35.3%) involved in cancer-related pathways (Figure 3C). Seven genes were associated with BC (APC, BRCA1, RB1, ESR1, ESR2, PTEN and PIK3CA); among these genes, only RB1 participates in cell cycle control, while the other six participate in different signalling pathways. The PI3K–Akt, Prolactin and oestrogen signalling pathways had the most common genes associated with BC, while the Hippo, Wnt and p53 signalling pathways shared one gene in common with BC (Table 2). Genome-wide methylation-associated biomarkers were involved in the pathways of neurodegeneration-multiple diseases (13%), cancer-related pathways (DCC, RASSF1, WNT1, CTNNA2; 13%), and Hippo, calcium signalling, cell adhesion and immune system-related pathways (all 10%) (Figure 3D).
Table 2. Signalling pathways and associated hypermethylation biomarkers
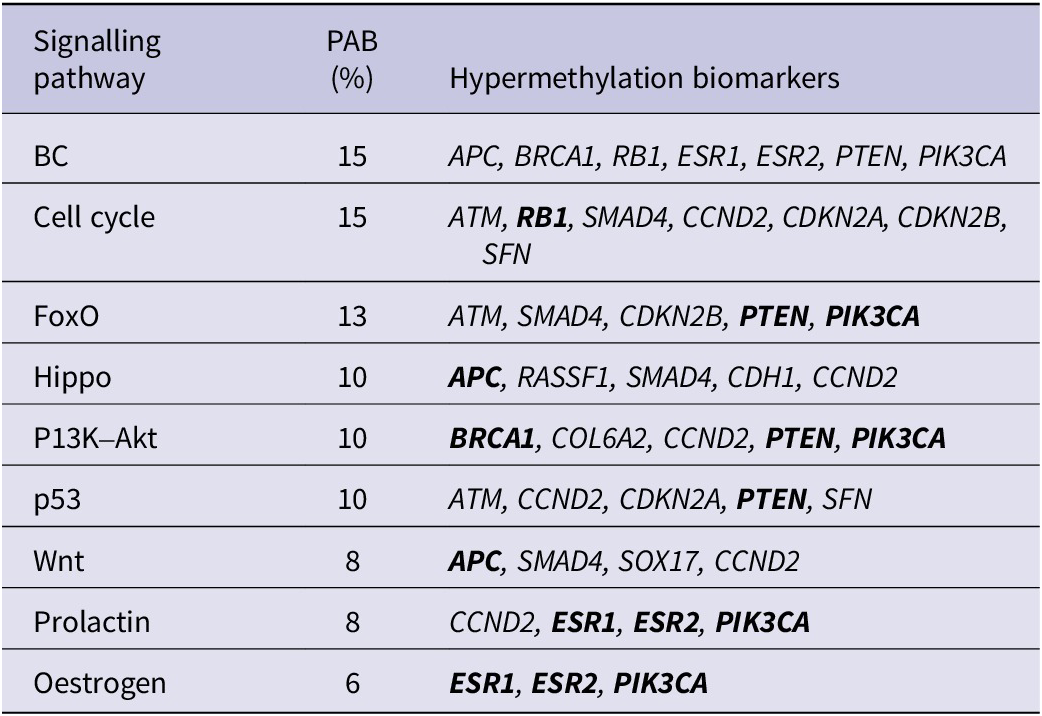
%: the percentage of the Set 1 genes associated with indicated signalling pathways; BC, breast cancer; PAB: pathway-associated biomarkers.
Note: The biomarkers in bold overlap with BC-associated biomarkers.
When data from methylome analysis with targeted biomarkers were compared, two BC-associated overlapping genes (SOX17 and RASSF1) were found. The main pathways of methylome biomarkers were associated with calcium signalling (CACNA1E, RYR2, RYR3), cancer pathways (DCC, RASSF1, WNT1, CTNNA2), multiple neurodegenerative diseases (WNT1, DNAI1, RYR2, RYR3), immune system disorders and cell adhesion (HLA-DRA, HLA-DRB1, HLA-DRB5).
Discussion
Current liquid biopsy utility in clinical practice
Liquid-biopsy assays analysing CTCs in BC have been in use in the clinic for nearly 20 years (Ref. Reference Hayes, Cristofanilli, Budd, Ellis, Stopeck, Miller, Matera, Allard, Doyle and Terstappen86). However, the only commercially available DNA methylation-based test for BC is the Therascreen® (QIAGEN) test (Ref. 87). The test is based on PITX2 (paired-like homeodomain transcription factor 2) methylation analysis in tumour tissue and is used to predict responses to anthracycline-based chemotherapy in ER-positive HER2-negative BC (Ref. Reference Schricker, Napieralski, Noske, Piednoir, Manner, Schüren, Lauber, Perkins, Magdolen, Schmitt, Ulm, Weichert, Kiechle, Martens and Wilhelm88). The test uses a Qiagen real-time PCR kit and runs in a Rotor-Gene Q MDx thermal cycler (QIAGEN) enabling the detection of three CpG sites in the PITX2 gene. DNA methylation of PITX2 predicts a poor response to chemotherapy in patients with lymph node-positive BC and the risk of distant metastasis in patients with node-negative BC (Refs Reference Maier, Nimmrich, Koenig, Eppenberger-Castori, Bohlmann, Paradiso, Spyratos, Thomssen, Mueller, Nährig, Schittulli, Kates, Lesche, Schwope, Kluth, Marx, Martens, Foekens, Schmitt and Harbeck89, Reference Harbeck, Nimmrich, Hartmann, Ross, Cufer, Grützmann, Kristiansen, Paradiso, Hartmann, Margossian, Martens, Schwope, Lukas, Müller, Milde-Langosch, Nährig, Foekens, Maier, Schmitt and Lesche90). Unfortunately, there is still no alternative test based on liquid biopsy.
Diagnostic and prognostic value of methylation biomarkers in liquid biopsies for BC
Our review analysis shows that certain methylation biomarkers present strong diagnostic and prognostic potential in BC, especially in more advanced disease settings. This review confirms that DNA methylation biomarkers analysed in liquid biopsies have strong potential to improve early BC detection and assist in the personalisation of treatment. Methylation analysis of genes such as SMAD4, PTEN, APC and RARB is suitable for early diagnosis and discrimination of malignant cases from non-cancerous controls with high sensitivity (>90%) and specificity (100%) and markedly outperforms clinically used protein biomarkers CEA and CA15.3. Some of the reviewed biomarkers have strong prognostic potential, allowing earlier determination of disease progression (APC, RASSF1, ESR1, TMEM240), association with BC stage (GBP2, RASSF1, APC, PTEN, SMAD4), lymph node metastasis (GBP2, ESR1, PTEN) or poor differentiation grade (ESR1, RASSF1, SMAD4). Moreover, ESR1, PTEN and TMEM240 methylation in liquid biopsy was associated with poor PFS and OS (Supplementary Table S1). In addition to specific genes, hypermethylation of the EFC#93 region, identified in an epigenome-wide study, was shown to predict the RFS and OS of patients with fatal BC 1 year before the event occurred (Ref. Reference Widschwendter, Evans, Jones, Ghazali, Reisel, Ryan, Gentry-Maharaj, Zikan, Cibula, Eichner, Alunni-Fabbroni, Koch, Janni, Paprotka, Wittenberger, Menon, Wahl, Rack and Lempiäinen52). Additionally, methylation of BC-related genes could provide essential insights into treatment response prediction, allowing clinicians to personalise treatment options according to the needs of individual patients, as shown by research investigating APC, BRCA1, PTEN, TMEM240, ENPP2 and ESR1 methylation profiles (Supplementary Table S1).
When analysing genes strongly associated with relevant carcinogenic pathways, several genes with essential functions were highlighted, including ESR1, PTEN, APC, SMAD4 and RASSF1. The genes RASSF1, SOX17 and DCC showed diagnostic potential in gene-targeted and epigenome-wide studies (Supplementary Tables S1 and S2). Moreover, the RASSF1 and SOX17 genes were overlapping in targeted and genome-wide analysis of our review analysis. Both genes are implicated in the regulation of cellular processes such as proliferation, differentiation and progression, and their hypermethylation has been associated with BC (Refs Reference Fu, Wang, Li-Chen, Wang, Shen, Huang and Shao91, Reference Gupta, Agarwal and Deshpande92). However, no overlap between the genomic regions selected for validation in various genome-wide studies was observed, pointing to the need for further technology development for bisulphite conversion and NGS of cfDNA fragments in liquid biopsy.
Based on this review, we can expect that testing for RASSF1, RARB, SMAD4, TMEM240, ESR1, PTEN, APC genes, and EFC#93 region hypermethylation can provide the most value due to their high diagnostic and prognostic sensitivity (Figure 4). Moreover, this set of genes is essential in the cell cycle, apoptosis, transcriptional regulation and cell migration processes and participates in various signalling pathways the dysregulation of which is crucial for cancer formation (Ref. Reference Feng, Spezia, Huang, Yuan, Zeng, Zhang, Ji, Liu, Huang, Luo, Liu, Lei, Du, Vuppalapati, Luu, Haydon, He and Ren93). It indicates that these biomarkers should be validated in independent BC cohorts.
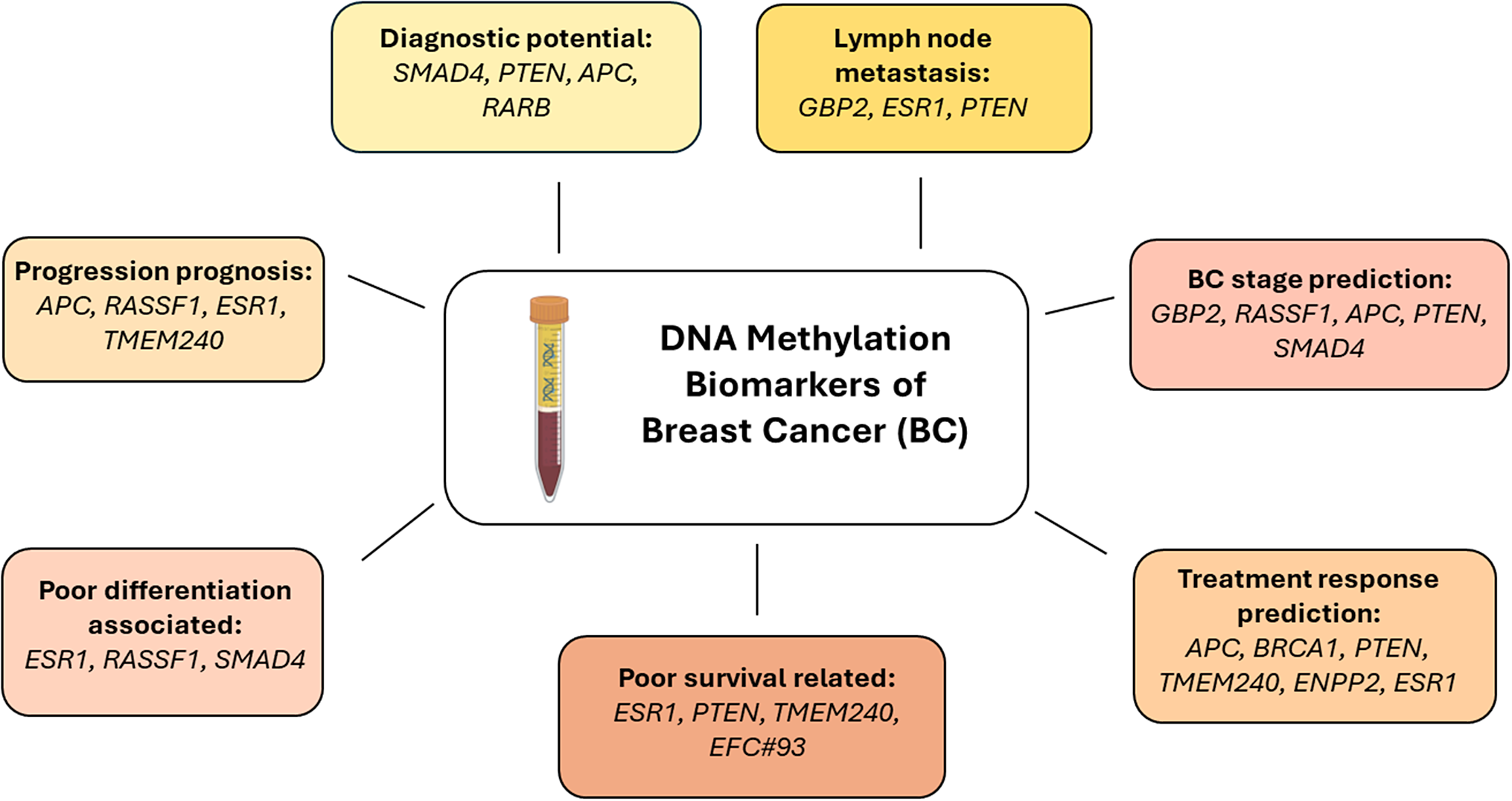
Figure 4. Breast cancer liquid biopsy biomarkers with the greatest diagnostic, prognostic or predictive significance.
Single-gene or gene set analysis by real-time or digital PCR techniques allows faster translation of laboratory-derived tests into in vitro diagnostics (IVD) tools. Such IVD tests are already available for colon (Cologuard® stool-DNA-based test; Epi proColon® 2.0 test; EarlyTect® CRC assay), cervical (Cervi-M® assay), lung (Epi proLung BL Reflex Assay®), glioblastoma (Therascreen MGMT Pyro Kit), bladder (Bladder EpiCheck®) and other tumours (Refs Reference Mancini, Righetto, Zumerle, Montopoli and Zattoni94, Reference Beltrán-García, Osca-Verdegal, Mena-Mollá and García-Giménez95). However, strict EU regulations and differences in national rules on clinical use and reimbursement policies remain the major issues for the wider application of such tests in the clinic.
Conclusions
Individualised consideration of DNA methylation profiles of tumour in conjunction with other clinical features would allow clinicians to adopt personalised patient management plans, facilitate early diagnosis and accurate prognosis, tailor treatment and monitoring strategies to each patient, and implement personalised medicine. While additional evidence is required before liquid biopsy-based DNA methylation tests can be implemented in the clinic, we can cautiously assume that they will have a place in BC management.
Abbreviations
- AUC
-
area under the curve
- BC
-
breast cancer
- cfDNA
-
cell-free DNA
- cfMETH
-
a predictive score of cfDNA methylation
- CTC
-
circulating tumour cell
- ctDNA
-
circulating tumour DNA
- DMCpGs
-
differentially methylated CpGs
- DMRs
-
differentially methylated regions
- EM
-
end motif
- EXE
-
exemestane
- GAD
-
Gene–Disease Association Database
- IVD
-
in vitro diagnostics
- KEGG
-
Kyoto Encyclopedia of Genes and Genomes
- MD
-
methylation density
- miRNA
-
microRNA
- MR
-
methylation ratios
- NGS
-
next-generation sequencing
- OS
-
overall survival
- PFS
-
progression-free survival
- ncRNA
-
non-coding RNA
- RRMP
-
reduced representative methylome profiling
- SPOT-MAS
-
screening for the presence of tumours by DNA methylation and size
- TGF-β
-
transforming growth factor beta
- TNBC
-
triple-negative breast cancer
- WGBS
-
whole-genome bisulphite sequencing
Supplementary material
The supplementary material for this article can be found at http://doi.org/10.1017/erm.2025.10008.
Data availability statement
The datasets supporting the conclusions of this article are included within the article and in Supplementary Tables S1 and S2.
Author contribution
I.S. performed the literature search, drafted the manuscript’s main part and participated in bioinformatics analysis. D.K. participated in manuscript writing and bioinformatics analysis. S.J. designed, supervised, edited and revised the manuscript. All authors have read and agreed to the final version of the manuscript.








