Introduction
Members of the class Archiacanthocephala are parasites found in terrestrial mammals and birds and are distributed worldwide. The class is divided into four orders: Apororhynchida, Gigantorhynchida, Moniliformida, and Oligacanthorhynchida, each with a single family (Amin Reference Amin2013; Bullock Reference Bullock and Schmidt1969). Archiacanthocephalans have an indirect life cycle using insects, myriapods, or other arthropods as intermediate hosts and terrestrial mammals or birds as definitive hosts. However, the participation of paratenic hosts, such as amphibians, turtles, snakes, and lizards, are key in the transmission of some parasites (Kenedy 2006; Nickol and Crompton Reference Nickol and Crompton1985). Currently, the family Oligacanthorhynchidae Southwell and Macfine, 1925 is divided into 12 genera, and one of the most diverse groups within the family is Oncicola Travassos, 1916. This genus includes 24 species that are generalist parasites able to infect a broad spectrum of mammalian carnivorous hosts, such as marsupials, procyonids, felids, and canids, and are globally distributed (Amin Reference Amin2013; Machado Filho Reference Machado Filho1950; Petrochenko Reference Petrochenko1958; Yamaguti Reference Yamaguti1963). Of the 24 described species of Oncicola, 16 of them are distributed in the Americas, representing 66.6% of the biodiversity of the genus. However, few species have been sequenced, limiting the understanding of the systematics of the family Oligacanthorhynchidae (García-Varela and Nadler Reference García-Varela and Nadler2006; Gazi et al. Reference Gazi, Sultana, Min, Chul Park, García-Varela, Nadler and Park2012; Near et al. Reference Near, Garey and Nadler1998).
In the neotropical region of Mexico, adults of two species of Oncicola have been recorded on the coasts of the Pacific Ocean and Gulf of Mexico, including O. luehei (Travassos, Reference Travassos1917) obtained from the North American opossum (Didelphis virginiana, Kerr) in Veracruz state and O. spirula (Olfers, 1816) obtained from the white-nosed coati (Nasua narica L.) in Chiapas state, whereas cystacanths (larval form) have been identified as Oncicola sp. or O. luehei in at least four amphibian species (i.e., Similisca cyanostica Smith, Lithobates forreri Boulenger, L. vaillanti Brocchi, and Rhinella marina L.) (García-Prieto et al. Reference García-Prieto, García-Varela, Mendoza-Garfias and Pérez-Ponce de León2010; Ortega-Olivares et al. 2013).
During a survey of parasitic helminths in northern and southeastern Mexico, adult specimens of an acanthocephalan were recovered from the digestive tract of a white-nosed coati (N. narica), whereas cystacanths were recovered from the body cavities of Vaillant’s Frog (L. vaillanti) and the Rio Grande Leopard Frog (Lithobates berlandieri Baird). After a morphological examination of worms from both stages, the adults and cystacanths were identified as O. luehei. Therefore, the objectives of this study were to i) characterize morphologically adults and cystacanths recovered from the intestines of white-nosed coatis and from the body cavities of their paratenic hosts (amphibians) from northern and southeastern Mexico; ii) link both stages of adults and cystacanths by using sequences of cytochrome c oxidase subunit 1 (cox1) from mitochondrial DNA; and iii) test the systematic position of O. luehei within Archiacanthocephala by using small (SSU) and large (LSU) subunits from nuclear ribosomal DNA.
Materials and methods
Sample collection
During several field expeditions in northern and southeastern Mexico, three common white-nosed coati (N. narica) were found in three localities: Chamela, Jalisco (19° 27’ 35.8’’ N, 104° 56’ 11.4’’ W), Ciudad Guzmán, Jalisco (19° 44’ 30. 934’’ N, 103° 28’ 29.33’’ W), and Catemaco, Veracruz (18° 26’ 14.43’’ N, 95° 04’ 52.387’’ W), and seven adult male Rio Grande Leopard Frogs (L. berlandieri) and eight adult female Vaillant’s Frogs (L. vaillanti) were collected in northern and southeastern Mexico (18º 35’–18 º 36’ N, 95 º 05’–95º 06’ W). The definitive and paratenic hosts were dissected, and the viscera were placed in separate Petri dishes with a 0.75% saline solution and examined under a dissecting microscope. The acanthocephalans were removed from the intestine (adult stage) and from the body cavity (encysted cystacanths) and washed in a 0.75% saline solution. Later, the unencysted cystacanths were placed in distilled water at 4°C overnight and subsequently were fixed and preserved in 70 or 100% ethanol.
Morphological analyses
A few acanthocephalans were gently punctured with a fine needle, stained with Mayer’s paracarmine, destained in 70% acid ethanol, dehydrated in a graded ethanol series, cleared in methyl salicylate, and mounted on permanent slides with Canada balsam. Each slide with a cystacanth was deposited in the Colección Nacional de Helmintos (CNHE), Instituto de Biología, Universidad Nacional Autónoma de México, Mexico City, under numbers 12226–12228.
The acanthocephalans were analysed with a Leica DM 1000 LED microscope equipped with bright field (Leica, Wetzlar, Germany). Acanthocephalans were initially identified by conventional morphological criteria following the key of Yamaguti (Reference Yamaguti1963) and the description of Machado Filho (Reference Machado Filho1950). For scanning electron microscopy (SEM), two adults were individually dehydrated with an ethanol series, critical point dried with CO2, sputter coated with gold, and examined with a Hitachi Stereoscan Model SU1510 scanning electron microscope operating at 15 kV at the Instituto de Biología, Universidad Nacional Autónoma de México (UNAM).
DNA sequence generation
A total of seven specimens identified as O. luehei were analyzed. Before DNA extraction, a tissue fragment was cut from two adults from northern and southeastern Mexico and two cystacanths (hologenophores, Pleijel et al. Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008), whereas the rest of the body was stained with Mayer’s paracarmine and mounted on permanent slides with Canada balsam. The tissue of each specimen was placed individually in tubes and digested overnight at 56°C in a solution containing 20 mM NaCl, 100 mM Na2 EDTA (pH 8.0), 10 mM Tris–HCl (pH 7.6), 1% sarkosyl, and 0.1 mg/ml proteinase K. Following digestion, genomic DNA was extracted from the supernatant using the DNAzol reagent (Molecular Research Center, Cincinnati, OH, USA) according to the manufacturer’s instructions. A fragment of the cytochrome c oxidase subunit 1 (cox 1) from the mitochondrial DNA was amplified using the forward primer 5′-AGTTCTAATCATAA(R)GATAT(Y)GG-3′ and reverse primer 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′ (Folmer et al. Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994). PCR amplifications were performed in a total volume of 25 μl containing 2 μl of each primer, 10 pmol/ μl, 2.5 μl of 10X buffer, 1.5 μl of 2 mM MgCl2, 2 μl of the genomic DNA, and 1U of Taq DNA polymerase (Platinum Taq, Invitrogen Corporation, California, United States). PCR cycling parameters for rDNA amplifications included denaturation at 94°C for 3 min, followed by 35 cycles of 94°C for 1 min, annealing at 40°C for 1 min, and extension at 72°C for 1 min, followed by a post-amplification incubation at 72°C for 7 min. Sequencing reactions were performed with the primers mentioned above using ABI Big Dye (Applied Biosystems, Boston, Massachusetts) terminator sequencing chemistry. Reaction products were separated and detected using an ABI 3730 capillary DNA sequencer. Contigs were assembled and base-calling differences were resolved using Codoncode Aligner version 11.0 (Codoncode Corporation, Dedham, Massachusetts).
Alignments, phylogenetic analyses, and haplotype network
Newly generated sequences cox 1, were aligned with published sequences for other members of Archiacanthocephala retrieved from the GenBank dataset (Table 1). Additionally, sequences from two nuclear genes from the SSU and LSU from Archiacanthocephalans were download from GenBank (Table 1) to test the systematic position of Oncicola. Alignments for each dataset (cox 1, SSU, and LSU) were constructed using the software Clustal W (Thompson et al. Reference Thompson, Higgins and Gibson1994). A nucleotide substitution model was selected for the dataset using jModelTest version 2.1.7 (Posada Reference Posada2008). Phylogenetic analyses were inferred through maximum likelihood (ML) with the program RAxML version 7.0.4 (Stamatakis Reference Stamatakis2006). A GTRGAMMAI substitution model was used, and 10,000 bootstrap replicates were run to assess nodal support. In addition, a Bayesian analysis was carried out, using the program MrBayes 3.2.2 (Ronquist et al. Reference Ronquist, Teslenko, Van Der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012) with two Markov chain Monte Carlo (MCMC) runs for 10 million generations, sampling every 1,000 generations, a heating parameter value of 0.2, and a burn-in of 25%. The resulting phylogenetics trees were visualized and edited using FigTree version 1.4.2 (Rambaut and Drummond Reference Rambaut and Drummond2007). Finally, uncorrected p distances were estimated with the cox 1 dataset by using the MEGA program (Kumar et al. Reference Kumar, Stecher and Tamura2016). To explore whether definitive and paratenic hosts from both coasts of Mexico share the same cox 1 haplotypes, an unrooted statistical network was constructed using PopART (Leigh and Bryant Reference Leigh and Bryant2015) with the minimum spanning network option (Bandelt et al. Reference Bandelt, Forster and Röhl1999).
Table 1. Classification and GenBank accession numbers of the specimens used in the phylogenetic analysis and haplotype network. Sequences in bold were generated in this study

Results
Morphological identification
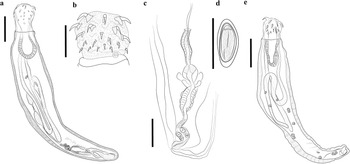
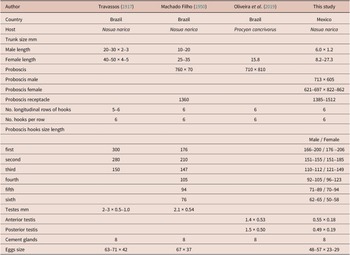
Adult acanthocephalans were recovered from intestines of three carcasses of white-nosed coati (N. narica) in Mexico (see Figure 1). The acanthocephalans showed similar morphological characteristics compared with those assigned to O. luehei by Travassos (Reference Travassos1917) and Machado Filho (Reference Machado Filho1950), including i) trunk cylindrical, narrow anteriorly enlarging midbody before tapering gradually to narrow posterior; ii) proboscis globular; iii) single-walled proboscis receptacle; iv) hooks in six alternative circles of six hooks each with roots robust, decreasing in size towards posterior; v) neck short with sensory papilla; vi) tubular lemnisci very long extending to the posterior region; vi) protonephridia dendritic type; vii) eight cement glands compact with single giant nuclei (Figures 2a–c). Compared to previous descriptions, our specimens exhibited variability in body size, proboscis, and hooks size (Table 2).

Figure 1. Sampling collection in Mexico. 1. Chamela, Jalisco (19° 27’ 35.8’’ N, 104° 56’ 11.4’’ W); 2. Ciudad Guzmán, Jalisco (19° 44’ 30. 934’’ N, 103° 28’ 29.33’’ W); 3. Catemaco, Veracruz (18° 26’ 14.43’’ N, 95° 04’ 52.387’’ W); 4. Los Tuxtlas, Veracruz (18º 35’–18 º 36’ N, 95 º 05’–95º 06’ W).

Figure 2. Drawing of Oncicola luehei from Nasua narica. Adult, total view (a); proboscis (b); Female reproductive system of Oncicola luehei (c); egg (d); cystacanth of Oncicola luehei from Lithobates vaillanti total view (e); Scale bars = 1.0 mm (a, e); 500 μm (b); 400 μm (c); 40 μm (d).
Table 2. Comparative metrical data for Oncicola luehei. Measurements in micrometres, unless otherwise indicated

Taxonomy
Class: Archiacanthocephala Meyer 1931
Order: Oligacanthorhynchida Petrochenko 1956
Family: Oligacanthorhynchidae Southwell and Macfie 1925
Genus: Oncicola Travassos 1916
Species: Oncicola luehei (Travassos Reference Travassos1917) Schmidt Reference Schmidt1972
Host: White-nosed coati Nasua narica Linnaeus
Site of infection: Intestine (prevalence 100%, (3/3)).
Paratenic host: Grande Leopard Frogs Lithobates berlandieri Baird; Vaillant’s Frogs Lithobates vaillanti Brocchi
Site of infection: Body cavity
Locality: Chamela, Jalisco (19° 27’ 35.8’’ N, 104° 56’ 11.4’’ W); Ciudad Guzmán, Jalisco (19° 44’ 30. 934’’ N, 103° 28’ 29.33’’ W); Catemaco, Veracruz (18° 26’ 14.43’’ N, 95° 04’ 52.387’’ W).
Voucher: CNHE No. 12226–228
Representative DNA sequences: PQ771482–488.
Redescription
General:
Sexual dimorphism evident, females larger than males. Trunk cylindrical, narrow anteriorly enlarging midbody before tapering gradually to narrow posterior (Figures 2a, 3a). Proboscis globular covered with 36 hooks in 6 circles of 6 hooks each (Figures 2b, 3b, 3c). Neck short with a sensory papilla (Figures 2b, 3d). Proboscis receptacle single-walled. Lemnisci elongate, reaching posterior end trunk. Protonephridia dendritic type. Gonopore subterminal in both sexes.

Figure 3. Scanning electron micrographs of adult specimen of Oncicola luehei, total view (a); proboscis (b, c); anterior region of proboscis (d). Arrows indicate a sensory papilla Scale bars = 1.0 mm (a); 500 μm (b); 400 μm (c, d).
Male (based on 1 mounted specimen). Trunk, 6.1 mm long × 1.2 wide. Proboscis 713 × 605, with 36 hooks in 6 alternative circles of 6 hooks each with roots robust, decreasing in size towards posterior (Figures 2b, 3a). The first hook row 166–200 (187), second 151–155 (153), third 110–112 (111), fourth 92–105 (99), fifth 71–89 (77), sixth 62–65 (63). Neck 347 × 653. Proboscis receptacle 1164 long. Lemnisci tubular very long extending near posterior end of trunk (Figure 2a). Testes ovoid in tandem, anterior 558 × 186, posterior 497 × 191. Eight cement glands compact with single giant nuclei (Figure 2a).
Female (based on 5 mounted specimens). 8.2–27.3 mm long (14.8 mm) × 2.3–4.1wide (2.9). Proboscis 621–697 (648) × 822–862 (848), with 36 hooks in 6 alternative circles of 6 hooks each with roots robust, decreasing in size towards posterior. The first hook row 176–206 (194), second 151–185 (168), third 121–149 (135), fourth 96–123 (105), fifth 70–94 (79), sixth 50–58 (54). Proboscis receptacle 1385–1512(1448) long. Female reproductive system short (Figure 2c). Gonopore subterminal. Mature eggs subspherical 48–57 (52), × 23–29 (26) (Figure 2d).
Cystacanth (Figure 2e) (based on two immature mounted specimens, (hologenophores)).
Trunk narrow anteriorly enlarging to widest point near midbody before tapering gradually to narrow posterior end; 6.1 mm (n=1) long by 1269 wide. Proboscis globular, 588–471 (529) long by 716–504 (610) wide; with 36 hooks in 6 alternative circles of 6 hooks each with roots robust, decreasing in size towards posterior. The first hook row 208–232 (224), second 178–207 (193), third 122–125 (123), fourth 66–92 (78), fifth 61–66 (63), sixth 56–60 (58). Neck 148–276 (212) long by 344–626 (485) wide; with pair of lateral sensory pits 47–54 (50) long by 44–67 (55) just posterior to root of last proboscis hook. Proboscis receptacle 1110 long by 522 wide (n=1) single walled with ventral cleft. Lemnisci tubular very long extending near posterior end of trunk, with small nuclei. Reproductive system primordial. Protonephridia dendritic type. Genital pore subterminal (see Figure 2e).
Phylogenetic analyses and haplotype network
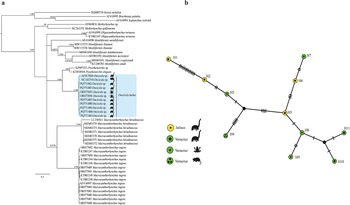
The cox 1 dataset included 664 sites and 49 sequences, and the best model was GTR + G + I. The tree inferred from the cox 1 dataset showed that the phylogenetic relationships among the genera Oligacanthorhynchus Travassos, 1915, Macracanthorhynchus Travassos, 1917, Oncicola and Prosthenorchis Travassos, 1915 from Oligacanthorhynchidae are unresolved due a polytomy at the base of the tree (Figure 4a). Our phylogenetic trees showed that the seven newly isolates identified as O. luehei formed a subclade together with two isolates identified as Oncicola sp. and O. luehei downloaded from GenBank (AF41700, NC_102754), plus two other isolates identified as Oncicola sp. (ORO77693-694) recovered from the body cavities of two amphibian species (Vaillant’s Frog (L. vaillanti) and Rio Grande Leopard Frog (L. berlandieri)) in southeastern Mexico (Figure 4a).

Figure 4. Phylogenetic trees using maximum likelihood (ML) and consensus Bayesian Inference for the cox 1 dataset (a). Numbers near internal nodes show ML bootstrap percentage values/ Bayesian posterior probabilities. Median-joining network of samples of Oncicola luehei built with the cox 1 gene (b). Each circle represents a haplotype, with size proportional to the haplotype’s frequency in the populations.
The uncorrected genetic divergence estimated with the cox 1 dataset between the clade formed by the isolates of Oncicola, and Prosthenorchis its sister taxa in the cox 1 phylogenetic tree, ranged 18%. The genetic divergence among our specimens of O. luehei recovered from three white-nosed coatis (N. narica) from northern and southeastern Mexico ranged from 0.03 to 1.5%; among the isolates recovered from the body cavities of their paratenic hosts, Vaillant’s Frog, and Rio Grande Leopard Frog ranged from 0 to 3%. Finally, the genetic divergence between the two isolates identified as Oncicola sp. and O. luehei (GenBank: AF41700; NC_102754) ranged 2%. Monophyly and low genetic distances among sequences of Oncicola sp. and O. luehei from northern and southeastern Mexico suggest that all the isolates belong same species.
The haplotype network built in this study was inferred with 11 specimens and 661 characters. The network inferred herein recognized 11 haplotypes. The haplotypes were separated from each other by a maximum of 13 substitutions. The haplotype network did not show a phylogeographic structure; therefore, the haplotypes could not be grouped into their own geographic clusters (Figure 4b).
To test the systematic position of O. luehei, within Oligacanthorhynchidae, two nuclear genes from the SSU and LSU were analysed. A previous sequence identified as Oncicola sp. (AF064818) by García-Varela et al. (Reference García-Varela, Pérez-Ponce de León, de la Torre, Cummings, Sarma and Laclette2000), now identified as O. luehei, was aligned together with 16 published SSU sequences from 12 species, plus four species from phylum Rotifera that were used as outgroup (see Table 1). The alignment contained 1,842 sites with 20 sequences. The best evolution model was TIM +I+G. This dataset included sequences representing four genera from Oligacanthorhynchidae:
Macracanthorhynchus, Oligacanthorhynchus, Pachysentis Meyer 1931, and Oncicola
The phylogenetic trees inferred with the SSU showed that Oligacanthorhynchidae is monophyletic and Oligacanthorhynchus is sister to Oncicola (Figure 5a). The LSU sequences of O. luehei (AY210467; AY829089) downloaded from GenBank were aligned together with 13 published sequences from eight species, plus four species from phylum Rotifera that were used as outgroup (see Table 1). The alignment contained 2,894 sites with 19 sequences. The best evolution model was GTR+G+I. This dataset included sequences representing three genera from Oligacanthorhynchidae; Macracanthorhynchus, Oligacanthorhynchus, and Oncicola. The topologies inferred with the LSU dataset agree with the SSU tree because both trees placed all the genera from Oligacanthorhynchidae in a clade. However, the sequences representing two species from Oncicola – O. luehei (AY210467, AY829089) and O. venezuelensis Marteau, 1977 (MK377340-341; KU521567) – were nested in two independent subclades, suggesting that the genus is paraphyletic (Figure 5b).

Figure 5. Phylogenetic trees using maximum likelihood (ML) and consensus Bayesian. SSU dataset (a), and LSU dataset (b). Numbers near internal nodes show ML bootstrap percentages/ Bayesian posterior probabilities.
Discussion
The acanthocephalan O. luehei obtained from the intestine of a white-nosed coati (N. narica) in Brazil was originally described as belonging to the genus Prosthenorchis (Travassoss, 1916). Later, it was validated in Yamaguti’s key (Yamaguti Reference Yamaguti1963). In Schmidt (Reference Schmidt1972), its designation in the class Archiacanthocephala was revised, a taxonomic rearrangement at the genus level was proposed, and several species initially classified as Prosthenorchis were transferred to Oncicola, including O. luehei, which was later recognized in Amin’s key (Amin Reference Amin2013). This acanthocephalan is a parasite of American carnivorous and insectivorous mammals since it has been recorded in the ring-tailed coati (Nasua nasua L), the crab-eating raccoon (Procyon cancrivorus Cuvier), Tate’s woolly mouse opossum (Marmosa paraguaya Tate), and the North American opossum (D. virginiana) in several countries from South America (Brazil, Argentina, and Paraguay) to North America (Mexico). In the present study, adult samples were collected from three white-nosed coatis (N. narica) in northern and southeastern Mexico, representing new records of this acanthocephalan in Mexico and extending its geographic range from northern Mexico to Argentina (Benatti et al. Reference Benatti, Moraes, Pacheco, Machado, Oliveira, Perin, Andrietti, Cândido Júnior, Vogliott, Tebaldi and Lux Hoppe2023; García-Prieto et al. Reference García-Prieto, García-Varela, Mendoza-Garfias and Pérez-Ponce de León2010; Hernández-Orts et al. Reference Hernández-Orts, Kutcha, Semenas, Cresco, González and Aznar2019; Oliveira et al. Reference Oliveira, Quagliatto Santos, Viotto de Souza, Iannini Custódio, Guilherme Lux-Hoppe and Rosalinski-Morales2019; Vieira et al. Reference Vieira, Luque and Muniz-Pereira2008). In addition, Ortega-Olivares et al. (Reference Ortega-Olivares, Velázquez-Urrieta, Sereno-Uribe, Harvey and García-Varela2023) reported that cystacanths were recovered from two frog species from southeastern Mexico. The cystacanths were initially identified as Oncicola sp.; morphologically, the specimens had a subglobular proboscis, covered with six hook rows with six hooks per row, and the lemnisci extended to the posterior region with small nuclei (Figure 2a–b). Nickol et al. (Reference Nickol, Fuller and Rock2006) reported that cystacanths from six genera of the family Oligacanthorhynchidae are characterized by having 36 proboscis hooks (Macracanthorhynchus, Oligacanthorhynchus, Oncicola, Prosthenorchis, and Tchadorhynchus Troncy, 1970). These genera can be distinguished at the species level primarily based on the features of the adult worms or molecular data (see Ortega-Olivares et al. Reference Ortega-Olivares, Velázquez-Urrieta, Sereno-Uribe, Harvey and García-Varela2023). In addition, the phylogenetic analyses of the cox 1 dataset confirmed that the sequences of Oncicola sp. (ORO77693-694) from two cystacanths (hologenophores) from southeastern Mexico formed a clade together with two other sequences available in GenBank (AF41700 and NC_102754) identified as Oncicola sp. and O. luehei, respectively, plus the new sequences from adult specimens identified as O. luehei from northern and southeastern Mexico, confirming that the specimens are conspecific. The intraspecific genetic divergence among the isolates of O. luehei from northern and southeastern Mexico was very low, ranging from 0 to 3%. The level of intraspecific genetic variation found is similar to that reported in other archicanthocephalans. For example, the genetic divergence among four isolates of Mediorhynchus gallinarum (Bhalerao 1937), a parasite of birds in Asia, was 0.2% (Rodríguez et al. Reference Rodríguez, Amin, Heckmann, Sharifdini and D’Elía2022); among 37 isolates from Prosthenorchis elegans (Diesing 1815), a parasite of New World primates and carnivores in South America, the intraspecific genetic divergence ranged from 0 to 1.6% (Falla et al. Reference Falla, Brieva and Bloor2015). Finally, among 15 isolates from Macracanthorhynchus ingens (von Linstow, 1879) Meyer 1932, a parasite of carnivores in North America, the intraspecific genetic divergence ranged from 0 to 2% (Ortega-Olivares et al. Reference Ortega-Olivares, Velázquez-Urrieta, Sereno-Uribe, Harvey and García-Varela2023).
Furthermore, the haplotype network analysis of cox 1 revealed 11 distinct haplotypes obtained from 11 individual sequences. Therefore, the haplotypes could not be grouped into geographic clusters. The lack of shared haplotypes between populations in northern and southern Mexico suggested a reduced recombination rate and a high pattern of genetic variation among the specimens, possibly because the collection sites are separated by mountains that form geographical barriers, including Sierra Madre del Sur, Sierra Madre Occidental, Sierra Madre Oriental, and the central Trans-Mexicana Volcanic Belt (Morrone et al. Reference Morrone, Escalante and Rodríguez-Tapia2017).
The cystacanths of O. luehei were found in two amphibian species (Vaillant’s Frog and Rio Grande Leopard Frog), which serve as paratenic hosts. Although the complete life cycle of O. luehei is unknown, the current evidence suggests that adult worms of the genus Oncicola live and reproduce sexually in the digestive tracts of carnivorous hosts (Kennedy Reference Kennedy2006). Female worms release eggs that are expelled into the environment with the faeces of the host. After the eggs are ingested by a termite that serves as an intermediate host, the parasite develops into the juvenile or cystacanth stage, at which point it is subsequently eaten by amphibians, lizards, and birds that serve as paratenic hosts until they are finally eaten by the appropriate definitive hosts (Nickol et al. Reference Nickol, Fuller and Rock2006).
The taxonomic history of the species of Oncicola have been unstable in that some of them were initially described in the genus Prosthenorchis and were subsequently transferred to the genus Oncicola (Schmidt, Reference Schmidt1972). The morphological traits that distinguish the two genera are the absence of a collar in Oncicola compared to the presence of a conspicuous festooned collar in the anterior region of the trunk in Prosthenorchis. The phylogenetic analyses based on the analysis of cox 1 placed Prosthenorchis spp. as a sister taxon to O. luehei (see Figure 4a), supporting the close relationships between the two genera. However, the LSU tree placed the two species of Oncicola analysed herein (O. luehei and O. venezuelensis) in two independent clades (Figure 5b), suggesting that the genus is paraphyletic. Morphologically, Oncicola is divided into two groups: the first has roughly saccular or pyriform-shaped trunks that are less than 20 mm long, similar to O. venezuelensis, while the second has elongated trunks that may reach lengths of nearly 50 mm, similar to O. luehei. In addition, the protonephridial organ is key to identifying species of Oncicola. The feature has not been described in O. venezuelensis, whereas O. luehei possesses a protonephridial organ, type dendritic, which has been observed in both adults and cystacanths (Nickol and Dunagan Reference Nickol and Dunagan1989; Schmidt Reference Schmidt1972).
Acknowledgements
We are grateful to Laura Marquez and Nelly López for their help during the sequencing of the DNA fragments. We also thank Berenit Mendoza Garfias for her help in obtaining the scanning electron microphotographs.
Author contribution
ALSU and MGV conceived and designed the study. ALSU, MTGG, MPOO, MIGM, and MGV wrote and edited the article. ALSU, MPOO, and MGV collected the samples. ALSU, MPOO, MTGG, and MGV designed the methodology.
Financial support
This research was supported by the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT-UNAM) IN201122.
Competing interest
No conflict of interest exists among the authors.
Consent to participate
All the listed authors have made significant contributions to the study and agreed to participate.
Consent for publication
All authors have read and agreed to the published version of the manuscript.
Ethical standards
The sampling in this work complies with the current laws and animal ethics regulations of México.