Introduction
The livestock sector of Pakistan comprises primarily farmhouses that supply meat, leather and milk (Ashraf et al., Reference Ashraf, Parveen, Asif, Awais, Khan, Aktas, Ozubek, Alanazi, Alyousif and Iqbal2020). Animals and their products are the main source of income in rural areas (Irshad et al., Reference Irshad, Qayyum, Hussain and Khan2010; Rehman et al., Reference Rehman, Nijhof, Sauter-Louis, Schauer, Staubach and Conraths2017a), and livestock production represents 13.4% of GDP, the country’s largest agricultural sector (Rehman et al., Reference Rehman, Jingdong, Chandio and Hussain2017b). Livestock production is dominated by small farmers following traditional methods of production, with 94% of farms owning fewer than 10 cattle or buffaloes, with these farms representing 67% of Pakistan’s cattle holdings (Horst and Watkins, Reference Horst and Watkins2022). Infestation by ecto- and endoparasites substantially impacts livestock production in Pakistan (Sajid et al., Reference Sajid, Iqbal, Khan and Muhammad2008; Shahzad et al., Reference Shahzad, Noor, Ahmad, Munir, Sharif Saghar, Hassan Mushtaq, Ahmad, Akbar and Mehmood2013; Jabbar et al., Reference Jabbar, Abbas, Sandhu, Saddiqi, Qamar and Gasser2015). For example, a study in Faisalabad found that tick infestation of cattle resulted in a morbidity rate of 43.7% and was the most prevalent disease condition reported (Ashfaq et al., Reference Ashfaq, Muhammad, Ul-Haq and Razzaq2014). Ticks also transmit many pathogens to animals and humans and contribute to the problem of livestock diseases (Parola and Raoult, Reference Parola and Raoult2001; Parola et al., Reference Parola, Paddock, Socolovschi, Labruna, Mediannikov, Kernif, Abdad, Stenos, Bitam, Fournier and Raoult2013; Luce-Fedrow et al., Reference Luce-Fedrow, Mullins, Kostik, St John, Jiang and Richards2015).
Tick-borne pathogens that affect humans are zoonotic in that they naturally circulate between ticks and vertebrate animal hosts with humans usually representing dead-end hosts. In Pakistan, most research into human tick-borne disease has focused on Crimean-Congo hemorrhagic fever virus (CCHFV), an endemic pathogen with a reported mortality rate as high as 40.7% in symptomatic cases (Umair et al., Reference Umair, Khurshid, Alam, Akhtar, Salman and Ikram2020). Although rickettsial pathogens that cause human disease are known to be present in Pakistan (Ullah et al., Reference Ullah, Alouffi, Almutairi, Islam, Rehman, Ul Islam, Ahmed, Júnior, Labruna, Tanaka and Ali2023), the distribution and diversity of these pathogens have been little studied representing a gap in the existing literature.
Rickettsia and Anaplasma (order Rickettsiales) are obligate intracellular bacteria. They are vector-borne pathogens mainly transmitted by ticks and cause diseases in humans and animals globally (Parola et al., Reference Parola, Paddock, Socolovschi, Labruna, Mediannikov, Kernif, Abdad, Stenos, Bitam, Fournier and Raoult2013). There are four major groups of genus Rickettsia and, among them, typhus group and spotted fever group (SFG) are associated with human diseases. Studies have recorded more than 25 species of SFG Rickettsia (Gillespie et al., Reference Gillespie, Williams, Shukla, Snyder, Nordberg, Ceraul, Dharmanolla, Rainey, Soneja, Shallom, Vishnubhat, Wattam, Purkayastha, Czar, Crasta, Setubal, Azad and Sobral2008; Thu et al., Reference Thu, Qiu, Matsuno, Kajihara, Mori-Kajihara, Omori, Monma, Chiba, Seto, Gokuden, Andoh, Oosako, Katakura, Takada, Sugimoto, Isoda and Nakao2019) (SFGR) with worldwide distribution. At present, 8 species of Anaplasma including A. marginale, A. phagocytophilum and A. ovis are recognized (Dumler et al., Reference Dumler, Barbet, Bekker, Dasch, Palmer, Ray, Rikihisa and Rurangirwa2001; Tate et al., Reference Tate, Howerth, Mead, Dugan, Luttrell, Sahora, Munderloh, Davidson and Yabsley2013; Li et al., Reference Li, Wang, Gao, Qi, Wang, Gao, Gao and Wang2015; Silaghi et al., Reference Silaghi, Santos, Gomes, Christova, Matei, Walder, Domingos, Bell-Sakyi, Sprong, von Loewenich, Oteo, de la Fuente and Dumler2017).
Anaplasmosis has been reported in wild and domestic ruminants and is caused by members of genus Anaplasma, which infect blood cells in animals. Anaplasma ovis and A. marginale cause pathogenicity in small and large ruminants, respectively (Kocan et al., Reference Kocan, de la Fuente, Blouin and Garcia-Garcia2004; Liu et al., Reference Liu, Ma, Wang, Wang, Peng, Li, Guan, Luo and Yin2012). Anaplasma ovis causes infections in sheep, deer and goats, whereas A. marginale causes anaplasmosis in cattle (Dumler et al., Reference Dumler, Barbet, Bekker, Dasch, Palmer, Ray, Rikihisa and Rurangirwa2001; Ashraf et al., Reference Ashraf, Khan, Khattak, Ali, Shaikh, Ali and Iqbal2013, Reference Ashraf, Parveen, Asif, Awais, Khan, Aktas, Ozubek, Alanazi, Alyousif and Iqbal2020; Seong et al., Reference Seong, Han, Chae, Chae, Yu, Lee, Park, Park, Yoo and Choi2015). Occasionally, A. ovis has also been reported as the causative agent of human infections (Chochlakis et al., Reference Chochlakis, Ioannou, Tselentis and Psaroulaki2010; Hosseini-Vasoukolaei et al., Reference Hosseini-Vasoukolaei, Oshaghi, Shayan, Vatandoost, Babamahmoudi, Yaghoobi-Ershadi, Telmadarraiy and Mohtarami2014). Tick species from the genera of Rhipicephalus, Dermacentor, Hyalomma, Ixodes and Argas have been reported to transmit Anaplasma spp. (Kocan et al., Reference Kocan, de la Fuente, Blouin and Garcia-Garcia2004; Hairgrove et al., Reference Hairgrove, Schroeder, Budke, Rodgers, Chung, Ueti and Bounpheng2015; Jabbar et al., Reference Jabbar, Abbas, Sandhu, Saddiqi, Qamar and Gasser2015; Battilani et al., Reference Battilani, De Arcangeli, Balboni and Dondi2017). Clinical findings of anaplasmosis in animals range from subclinical in < 1 year old to severe and often fatal in older cattle, and are characterized by anaemia, rapid loss of condition, reduced milk production, anorexia and abortion in pregnant animals (Aubry and Geale, Reference Aubry and Geale2011; Jaswal et al., Reference Jaswal, Bal, Singla, Gupta and Brar2015).
Pathogenic SFGR have been reported from ticks all over the world (Piotrowski and Rymaszewska, Reference Piotrowski and Rymaszewska2020; Zhang et al., Reference Zhang, Sun, Chen, Teng, Wang, Li, Hay, Fang, Yang and Liu2023), and they are among the oldest known vector-borne diseases (Parola et al., Reference Parola, Paddock and Raoult2005). SFGR have been insufficiently studied in Pakistan although previous studies have found that rickettsiae, including known pathogenic species, circulate in Pakistani ticks and livestock (Karim et al., Reference Karim, Budachetri, Mukherjee, Williams, Kausar, Hassan, Adamson, Dowd, Apanskevich, Arijo, Sindhu, Kakar, Khan, Ullah, Sajid, Ali and Iqbal2017; Ali et al., Reference Ali, Shehla, Zahid, Ullah, Zeb, Ahmed, da Silva Vaz and Tanaka2022a, Reference Ali, Numan, Khan, Aiman, Muñoz-Leal, Chitimia-Dobler, Labruna and Nijhof2022b; Zeb et al., Reference Zeb, Song, Khan, Senbill, Aziz, Hussain, Sánchez, Cabezas-Cruz, Alzahrani, Alshehri, Alghamdi and Sparagano2024). The aim of the present study was to improve understanding of the diversity, distribution and prevalence of SFGR and Anaplasma spp. in ticks from three provinces of Pakistan and to describe the phylogenetic relationships of these pathogens to existing records.
Materials and methods
Tick sampling and identification
Ticks from Punjab, Khyber Pakhtunkhwa and Islamabad were investigated. The majority of cattle, buffaloes and goats in the Pakistan are reared in Punjab and Khyber Pakhtunkhwa provinces (Pakistan Bureau of Statistics, 2006). In a previous study, ticks were collected from 1325 cattle (Bos indicus; B. taurus), 127 sheep (Ovis aries), 89 buffaloes (Bubalus bubalis) and 539 goats (Capra hircus; Khan et al., Reference Khan, Ahmed, Afzal, Khan, Birtles and Oliver2022). Of these, 1129 animals were infested with ticks. Ticks were preserved in 70% ethanol and male ticks were identified morphologically. In total, 390 individual male ticks representing 10 species were selected for DNA extraction and testing for the presence of Rickettsia and Anaplasma. Further details on tick species identification and animal infestation prevalence and risk factors can be found in (Khan et al., Reference Khan, Ahmed, Afzal, Khan, Birtles and Oliver2022).
DNA extraction
Specimens in ethanol were washed and rehydrated twice in 1x PBS for 20 min. To remove all the ethanol residues, they were dried for an additional 20 min. DNA was extracted from all ticks using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). Minor changes to the protocol were done according to the manufacturer instructions: proteinase K incubation was carried on overnight at 56 °C and DNA was eluted in two steps with 25 µL each of sterile water pre-heated to 72 °C. DNA was quantified and stored at −80 °C until use. The tick samples were pooled prior to sequencing to study the diversity of the microbial communities.
Nested PCR
A total of 390 ticks were screened for SFGR and Anaplasma infections. The presence of SFGR was determined using ompA gene-specific primers (Mukherjee et al., Reference Mukherjee, Beati, Sellers, Burton, Adamson, Robbins, Moore and Karim2014). For primary PCR, RR190-70 (50-ATGGCGAATATTTCTCCAAAA-30) and RR190-701 (50-GTTCCGTTAATGGCAGCATCT-30) primers were used; whereas for nested PCR, 190-FN1 (50-AAGCAATACAACAAGGTC-30) and 190-RN1 (50-TGACAGTTATTATACCTC-30) primers were used. 2.5 μL of DNA template was added to 8 μL of ultra-pure water, 12.5 μL of 2 × PCR Master Mix and 1 μL of each primer in the primary reaction. 2.5 μL of the primary PCR, 12.5 μL of 2 × PCR Master Mix, 1 μL of each nested primer and 8 μL of ultra-pure water were used in the nested reaction. PCRs were performed using the following sequence: 1 cycle at 95 °C for 3 min, 35 cycles at 95 °C for 20 sec, 46 °C for 30 sec and 63 °C for 60 s and 1 cycle at 72 °C for 7 min. DNA from Rickettsia parkeri was used as a positive control, and PCR mix with no DNA template was used as a negative control.
For detection of Anaplasma, we used EBR2, EHR 16SD and EBR3 primers. The EHR 16SD (5 – GGTACCYACAGAAGAAGTCC-3) primer amplified 16S rDNA from all members of the Anaplasmataceae as described by Hornok et al. (Reference Hornok, Földvári, Elek, Naranjo, Farkas and de la Fuente2008). EBR2 (5 -TGCTGACTTGACATCATCCC-3) and EBR3 (5 – TTGTAGTCGCCATTGTAGCAC-3) primers were described by Teshale et al. (Reference Teshale, Geysen, Ameni, Asfaw and Berkvens2015). We used EHR 16SD primer as a forward primer in the first round of amplification as it is already proved to be suitable for Anaplasma species determination and again used it for the second cycle of amplification as a forward external primer.
Nested PCR used EHR 16SD and EBR3 primers for the first round of PCR amplification to amplify a fragment of about 925 bp (16S rDNA gene). EHR 16SD was used as forward external primer, and EBR2 was used as internal primer for the second round of PCR amplification. Master mix was the same as used for ompA. The conditions for the first round were 92 °C for 3 min, 40 cycles of denaturation at 92 °C for 30 s, annealing for 45 s at 62 °C, elongation for 1 min at 72 °C and final extension for 10 min at 72 °C . PCR product (0.5 μL) from the first reaction was used as a template for the second round of amplification, which lasted for 25 cycles. Throughout the PCR procedures, negative samples were retested for any possible inhibition effect. DNA from A. phagocytophilum was used as a positive control, and PCR mix with no DNA template was used as a negative control.
DNA sequencing and phylogenetic analysis
All the PCR products were examined by gel electrophoresis on 2% agarose gel containing ethidium bromide. PCR products (540 bp in length for Rickettsia and 925 bp for Anaplasma) were excised from the agarose gel, and then DNA extraction was done using a Monarch DNA gel extraction kit (New England BioLabs, Ipswich, MA, USA). The purified DNA samples were sent to Genomic Centre of University of Minnesota for Sanger sequencing. Gene amplicons were subjected to BLAST analyses for verification of Rickettsia and Anaplasma identification.
For both Rickettsia and Anaplasma PCR amplicons, Sanger sequence data were generated using forward and reverse primer pairs. Sequences were aligned and manually inspected using Geneious Prime (v2021.2.2) (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Meintjes and Drummond2012). Pair-wise alignments were trimmed following visualization of each corresponding sequence chromatogram, and any sequences with poor chromatogram quality were excluded from downstream analyses. The resulting trimmed consensus sequences were aligned using MAAFT (v7.450) (Katoh and Standley, Reference Katoh and Standley2013).
Maximum likelihood phylogenies were generated using RAxML (v8.2.11) (Stamatakis, Reference Stamatakis2014) with the following parameters: rapid bootstrap analysis mode; 1000 bootstrap iterations; general time-reversible substitution model; and gamma model of rate heterogeneity (i.e. GTR + G). For the phylogenetic tree based on Rickettsia ompA gene, Rickettsia tamurae (AB114823) was used as an outgroup. Similarly, for the phylogeny based on Anaplasma 16S gene, Ehrlichia ruminantium (GenBank Accession no. DQ482915) was used as an outgroup. Phylogenetic trees were visualized and annotated using the Interactive Tree of Life online tool (v6.5.2, https://itol.embl.de/) (Letunic and Bork, Reference Letunic and Bork2024).
Results
Molecular detection of putative pathogenic spotted fever group of Rickettsia and Anaplasma
A total of 390 ticks collected from sheep, goats, cattle and buffaloes from different tick species, e.g. Haemaphysalis punctata, H. sulcata, Hyalomma anatolicum, Hy. dromedarii, Hy. excavatum, Hy. rufipes, Hy. scupense, Rhipicephalus decoloratus, Rh. microplus and Rh. sanguineus, were used in detection of SFGR and Anaplasma.
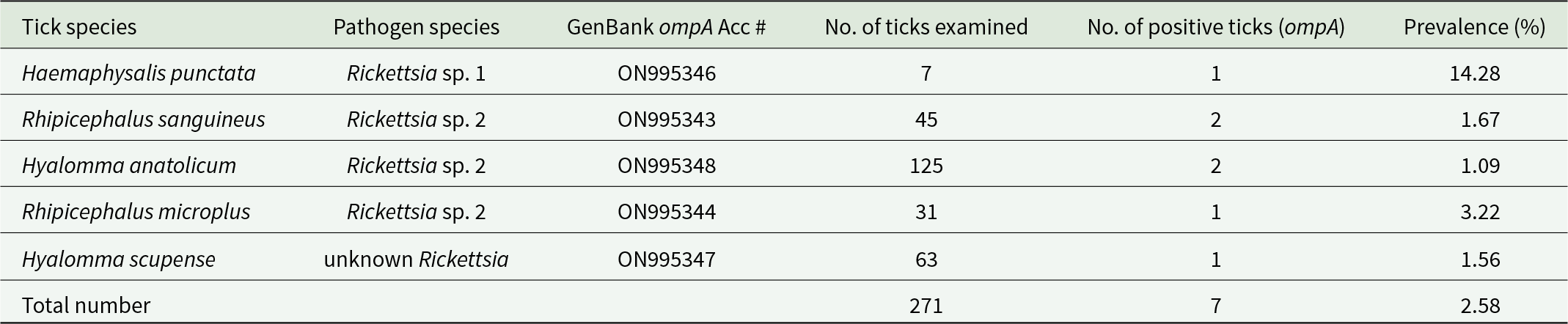
The RR190-70, RR190-701, 190-FN1 and 190-RN1 primers amplified parts of the rickettsial ompA gene. After amplification, bacterial DNA gave a clear single band at 540 bp. Positive samples comprised 7/390 (2.58%) belonging to 5 tick species. SFGR DNA was not amplified from Hy. dromedarii, H. sulcata and Hy. rufipes. According to nucleotide sequences, unknown Rickettsia sp. was detected in 14.28% (1/7) of H. punctata and uncultured Rickettsia sp. 2 was observed in 1.67% (1/60) of Rh. sanguineus, 1.09% (1/91) of Hy. anatolicum and 3.22% (1/31) of Rh. microplus. Rickettsia sp. 3 was detected in 1.56% (1/64) of Hy. scupense. Tick species positive SFGR and their prevalence are listed in Table 1.
Table 1. Putative pathogenic SFG Rickettsia (ompa) in tick species collected from variety of hosts
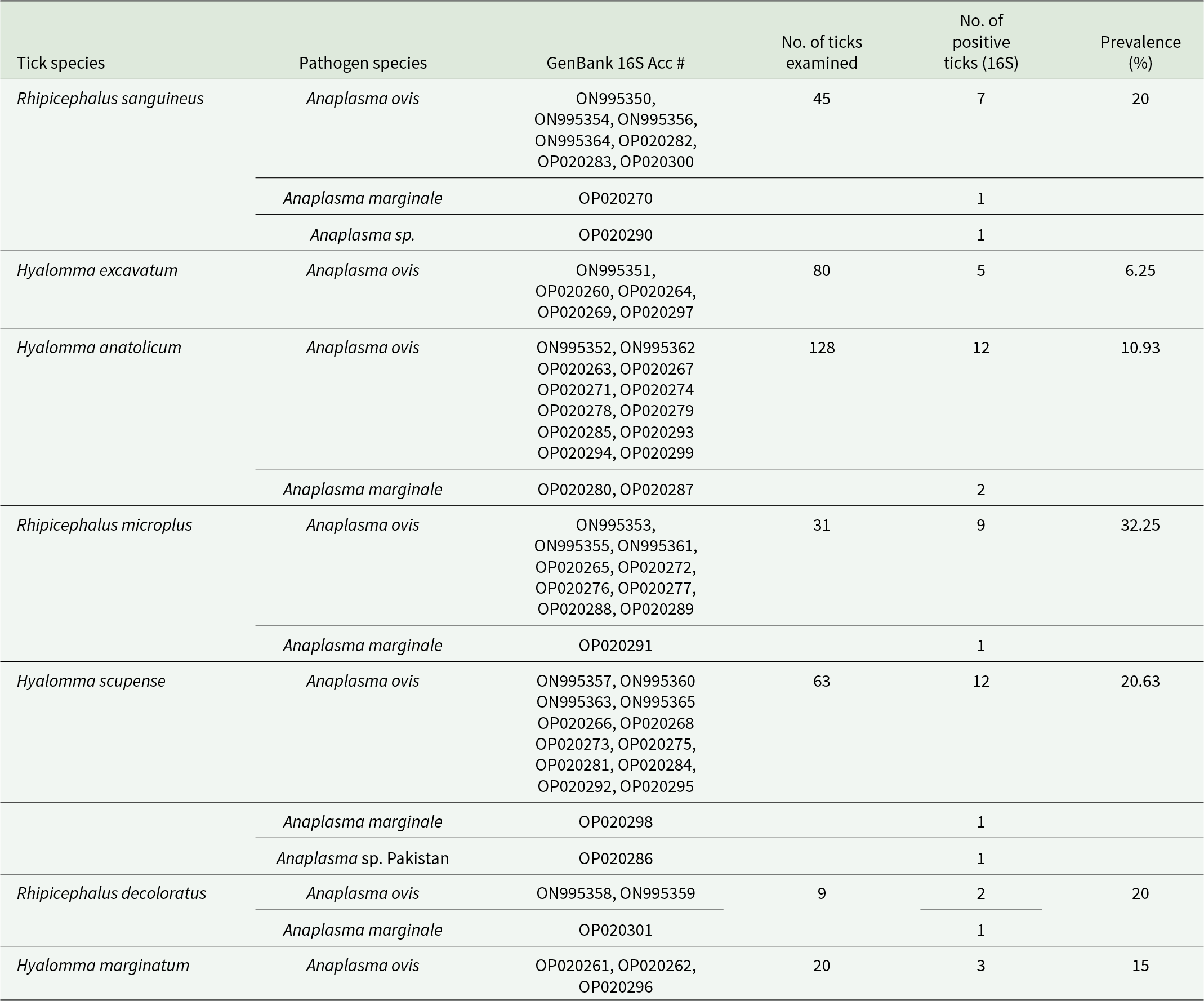
The EBR2, EBR3 and 16SD primers amplified parts of the Anaplasma 16S rDNA gene. After amplification, bacterial DNA gave a clear single band at 925 bp. Among the tested ticks, 57/390 (14.62%) were positive for Anaplasma spp. DNA isolated from Hy. dromedarii, H. punctata, H. sulcate and Hy. rufipes did not amplify Anaplasma. A. ovis and A. marginale were detected in 32.25% of Rh. microplus, 20.63% of Hy. scupense, 20% of Rh. sanguineus, 6.25% of Hy. excavatum and 10.93% of Hy. anatolicum (Table 2).
Table 2. Putative pathogenic anaplasma (16S) in tick species collected from variety of hosts
Phylogeny of bacteria
Phylogenetic tree based on rickettsia ompa gene
All tick sequences included in this study (GenBank accession nos ON995343–ON995349) formed three monophyletic clades. The first clade consists of two sequences (GenBank accession nos. ON995346 and ON995345) obtained from H. punctata and Rh. sanguineus, respectively, and are closely related to Rickettsia japonica and Rickettsia heilongjiangensis. A high bootstrap value of > 98 indicates that they belong to the same genotypes. Both of these sequences originated in China, and the hosts of both these samples were Haemaphysalis ticks.
The second cluster is formed by four sequences (GenBank accession nos. ON995343, ON995344, ON995348 and ON995349). Their strong relationship is supported by a high bootstrap value of 100. They are closely related to Rickettsia rickettsii (HM446486.1), Rickettsia philipii (EU109181) and Rickettsia honei (DQ309096). Rickettsia rickettsii (HM446486.1) and R. philipii (EU109181) were obtained from Ambylomma americanum and Dermacentor occidentalis, respectively, and the origin of both these sequences was the USA.
The sequence (GenBank accession no. ON995347) makes a distinct clade, supported by a high bootstrap value of 95. This sequence is genetically similar to the AB795207 sequence, originating from Uzbekistan. It also shares a close evolutionary relationship with another rickettsial sequence obtained from Madagascar from Ambylomma variegatum having GenBank accession no. KR492932.1. The tree is rooted by R. tamurae, which acts as an outgroup.
Phylogenetic tree based on Anaplasma 16S gene
The tree is well rooted by Ehrlichia ruminatum, which acts as an outgroup in this tree. The first ingroup, which is separated by a node having a bootstrap value of 100, is formed by A. phagocytophilum (GenBank accession nos. GU046565, AF481850 and AB196720). The ancestors for A. platys and A. bovis were recently separated from the cluster formed by the sequences included in this study. Tick sequences in this study formed two distinctive monophyletic clades with high bootstrap values. Both these sequences are grouped together, indicating they share similar genotypes. The first clade (GenBank accession nos. ON995350–ON995364) is genetically identical to A. ovis (EF587237) and A. centrale (AF309869) and share a common ancestor. Anaplasma ovis (EF587237) originated from China and is nestled between ON995350 and ON995364, while A. centrale (AF309869) is a vaccine strain obtained from Israel.
All the other 42 sequences from different tick species in this study (GenBank accession nos. OP020260–OP020301) were clustered together and phylogenetically related to A. marginale (GenBank accession nos. DQ341369 and DQ341370) from South China. Their close relation was supported statistically by a high bootstrap value of 99.
The phylogenetic trees of Rickettsia and Anaplasma are shown in Figure 1 and Figure 2.
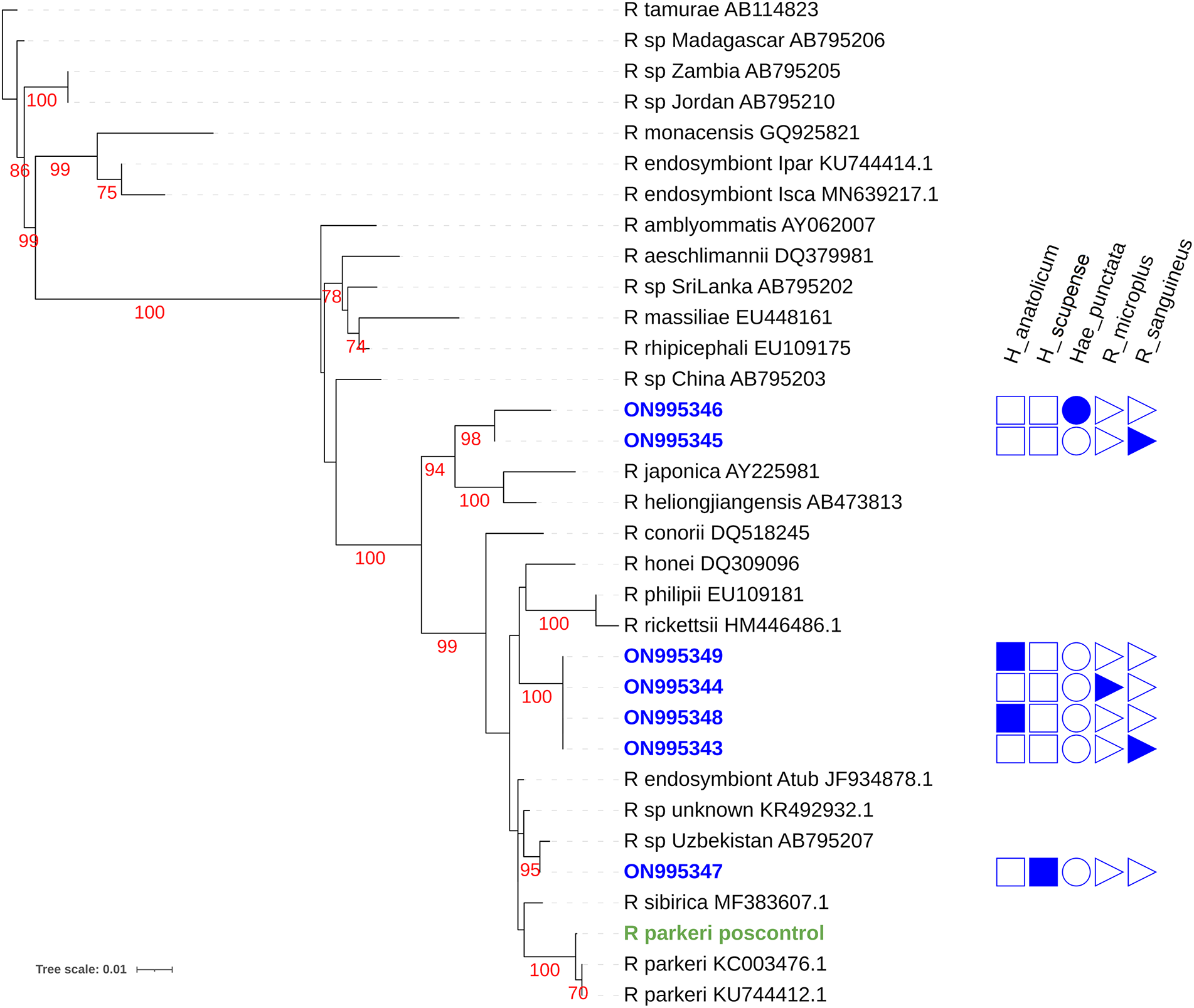
Figure 1. Maximum likelihood phylogenetic tree for the Rickettsia ompa gene for taxa detected in this study and others listed in NCBI genbank. Sequences obtained in this study are listed in blue as a genbank accession number with an indication of the tick species from which it was discovered. Red numbers indicate bootstrap values. Green text indicates the R. parkeri DNA positive control.

Figure 2. Maximum likelihood phylogenetic tree for the Anaplasma 16S rDNA gene for taxa detected in this study and others listed in NCBI genbank. Sequences obtained in this study are listed as a blue genbank accession number with an indication of the tick species from which it was discovered. Red numbers indicate bootstrap values. The green box indicates likely A. ovis sequences and the orange box indicates likely A. marginale sequences.
Discussion
SFGR are neglected diseases in developing countries across Asia and are emerging infections in all over the world (Chikeka and Dumler, Reference Chikeka and Dumler2015). The sub-tropical climate, humidity and high temperature result in the spread of ticks and tick-borne diseases in Pakistan, but the prevalence rates of these pathogens in ticks have not been much investigated (Ashraf et al., Reference Ashraf, Parveen, Asif, Awais, Khan, Aktas, Ozubek, Alanazi, Alyousif and Iqbal2020). It is likely that a changing climate and associated changes in temperature and moisture patterns will affect tick distributions, phenology and disease transmission risks in unpredictable ways, further emphasizing the need for ongoing surveillance efforts (de Souza and Weaver, Reference de Souza and Weaver2024).
SFGR and Anaplasma were detected by PCR amplification of ompA and 16S gene. These genes have been used in previous studies to detect the presence of these pathogens in ticks (Baldridge et al., Reference Baldridge, Burkhardt, Simser, Kurtti and Munderloh2004; Teshale et al., Reference Teshale, Geysen, Ameni, Asfaw and Berkvens2015). Based on the single gene phylogeny (Figure 1), potentially novel SFGR were found in Hy. scupense, H. punctata and R. sanguineus ticks. Given the understudied nature of rickettsiae in Pakistan and global diversity of the clade, finding SFGR of uncertain identity is not surprising. Phylogenetic placement provides no evidence of whether these represent potential human or livestock pathogens. Further investigation of rickettsial distribution in ticks and associated livestock is warranted. Producing more complete genetic data to better define the phylogenic positions of potentially undescribed SFGR would also be of value in predicting risk of pathogenicity and better understanding SFGR biodiversity.
This study reported Rickettsia spp. in ticks collected from livestock. A study conducted in Khyber Pakhtunkhwa, Pakistan tested ticks collected from livestock and found 20.4% positivity for Rickettsia (Ali et al., Reference Ali, Shehla, Zahid, Ullah, Zeb, Ahmed, da Silva Vaz and Tanaka2022a). Another study conducted in the Federally Administered Tribal Areas of Khyber Pakhtunkhwa, Pakistan, reported prevalence of Rickettsia massiliae (42.6%), Rickettsia slovaca (25.9%) and Rickettsia conorii (5.6%) in ticks collected from small ruminants (Ghafar et al., Reference Ghafar, Cabezas-Cruz, Galon, Obregon, Gasser, Moutailler and Jabbar2020), demonstrating that pathogenic SFGR are actively in circulation in Pakistan. In another study, SFGR-specific amplicons also were identified in 10% of ticks (514) collected from livestock, including from the potential pathogen Rickettsia amblyommatis (Karim et al., Reference Karim, Budachetri, Mukherjee, Williams, Kausar, Hassan, Adamson, Dowd, Apanskevich, Arijo, Sindhu, Kakar, Khan, Ullah, Sajid, Ali and Iqbal2017).
Our study found A. ovis and A. marginale in tick samples collected from buffaloes, cattle, sheep and goats. Many previous studies have reported on bovine anaplasmosis-associated pathogens in large ruminants but Anaplasma prevalence in small ruminants needs more investigation (Zabel and Agusto, Reference Zabel and Agusto2018). Abid et al. (Reference Abid, Bukhari, Asif, Sattar, Arshad, Aktas, Ozubek, Shaikh and Iqbal2021) conducted a study in Layyah based on the detection of the msp1b gene in sheep and reported 6.9% of sheep as positive for A. marginale. Ghaffar et al. (Reference Ghaffar, Ijaz, Ali, Farooqi, Rehman, Ali, Zafar and Naeem2020) did a study in Mianwali, Pakistan, targeting 16S rRNA gene and found many sheep (32%) positive for A. marginale and A. ovis. In other studies, 40% of sheep were positive for A. marginale on the basis of MSP5 indirect ELISA in Peshawar and 16.2% of sheep from Lakki Marwat and Peshawar were positive for A. marginale (Kashif and Ahmad, Reference Kashif and Ahmad2014; Turi, Reference Turi2018). Hussain et al. (Reference Hussain, Junaid, Gul, Jamal, Ahmed, Talpur, Rahim, Fatima and Munir2017) reported that 42.7% of sheep in Karak, Khyber Pakhtunkhwa were positive for A. marginale. Many factors may affect the prevalence of A. marginale in sheep including habitat, tick control programmes, abiotic factors and the management methods of livestock farms (Belkahia et al., Reference Belkahia, Ben Said, Alberti, Abdi, Issaoui, Hattab, Gharbi and Messadi2015).
The presence of SFGR and Anaplasma spp. in ticks collected from small ruminants represents a particular concern given the relative lack of research into this host/pathogen relationship. Sheep have been demonstrated to develop clinical illness when infected with A. marginale (Abdullah et al., Reference Abdullah, Ali, Jasim, Ola-Fadunsin, Gimba and Ali2020) and represent a likely reservoir for the infection of cattle. Further research is warranted to determine best practices in livestock husbandry to limit tick feeding on livestock, develop and distribute effective A. marginale vaccines to both cattle and small ruminants, and promote pasturage methods to limit contact between herds. More systematic surveillance of livestock infection, ticks and tick-borne pathogen diversity and biology will be required to accurately assess risk. Surveillance studies are particularly lacking in southern Pakistan where socioeconomic conditions are poorer and livestock rearing is more focused on smaller ruminants than on cattle. Improved surveillance, public education and research-informed policy development and enforcement will be needed to curb the spread of tick-borne livestock and human diseases in Pakistan.
Data availability statement
Sequencing data is available on National Center for Biotechnology Information GenBank (https://www.ncbi.nlm.nih.gov/genbank/).
Authors contributions
S.S.K., H.A. and J.D.O. conceived and designed the study. S.S.K. and B.K. performed experimentation and data collection. E.J.K. and M.U.A. performed phylogenetic analyses, S.S.K., H.A., A.A., D.S. and J.D.O. wrote and reviewed the article.
Financial support
This research received funding from Higher Education Commission of Pakistan.
Competing interests
We declare no conflicts of interest.
Ethical standards
The experimental design of this study was approved by the Ethical Committee of COMSATS University (permit no. CUI/Bio/ERB/2021/42). No livestock were harmed in the collection process.







