Introduction
Microbiomes in astrobiology
Astrobiology, the study of life in the universe, seeks to understand the origins, evolution, and potential distribution of life on early Earth and other celestial bodies. A critical aspect of this field is the investigation of microorganisms, particularly extremophiles, which thrive in harsh environments such as deep-sea vents, acidic hot springs, and polar ice caps (Rappaport and Oliverio, Reference Rappaport and Oliverio2023; Thakur et al., Reference Thakur, Singh and Zhang2022). These organisms provide analogs for potential extraterrestrial life forms, offering insights into how life might adapt to extreme conditions on other planets, such as Mars and Venus or Saturn’s and Jupiter’s moons like Europa or Enceladus (Limaye et al., Reference Limaye, Mogul, Smith, Ansari, Słowik and Vaishampayan2018; Weber et al., Reference Weber, Marlin, Prakash, Teece, Dzurilla and Barge2023).
Microbiome research has revolutionized our understanding of microbial diversity and adaptability. Next-generation sequencing technologies, such as 16S rRNA gene sequencing and metagenomics, have enabled the identification and characterization of microbial communities in previously inaccessible environments, including the atmosphere, by measuring air samples at different altitudes. These tools allow researchers to uncover microorganisms’ genetic and functional potential, even in low-biomass samples like those from high-altitude air masses (González-Martín et al., Reference González-Martín, Pérez-González, González-Toril, Expósito, Aguilera and Díaz2021; Jiang et al., Reference Jiang, Wang, Guo, Hou, Guo, Zhang, Tan, Li, Li and Zhu2022).
The Earth’s atmosphere is a dynamic and stratified ecosystem that supports microbial life under varying conditions. The troposphere, the lowest atmospheric layer, contains 75% of the atmosphere’s mass, most of its water vapor, and the majority of airborne microorganisms (Šantl-Temkiv et al., Reference Šantl-Temkiv, Amato, Casamayor, Lee and Pointing2022). This layer is characterized by turbulent mixing, weather phenomena, and the presence of particles that serve as vehicles for microbial dispersal. Studies have shown that microbes in the troposphere can travel thousands of kilometers via long-range air currents, often hitching rides on particles such as desert dust or marine aerosols (Rodó et al., Reference Rodó, Pozdniakova, Borràs, Matsuki, Tanimoto, Armengol, Pey, Vila, Muñoz, Santamaria, Cañas, Morguí, Fontal and Curcoll2024; Šantl-Temkiv et al., Reference Šantl-Temkiv, Amato, Casamayor, Lee and Pointing2022). This global dispersal mechanism plays a crucial role in shaping microbial diversity and influencing ecological interactions across continents. On the other hand, microbial diversity at higher altitudes decreases. With increased exposure to low temperature, UV radiation, and limitation of nutrients, higher altitude environments present more extreme conditions compared to lower parts of the atmosphere. Microbial survival there is primarily restricted to stress-tolerant taxa, such as spore-forming bacteria and fungi. While viable microbes have been detected at these altitudes, their abundance and diversity are significantly lower than in the troposphere due to prolonged exposure to harsh conditions (Šantl-Temkiv et al., Reference Šantl-Temkiv, Amato, Casamayor, Lee and Pointing2022; Smith et al., Reference Smith, Ravichandar, Jain, Griffin, Yu, Tan, Thissen, Lusby, Nicoll, Shedler, Martinez, Osorio, Lechniak, Choi, Sabino, Iverson, Chan, Jaing and McGrath2018).
Atmosphere microbiomes – aerobiomes
Aerobiome research focuses on airborne microbial communities and their ecological roles. Recent studies have revealed that microbes are not merely passively transported through the air but can remain metabolically active under certain conditions. For instance, Pseudomonas syringae found in clouds have been shown to influence precipitation patterns by acting as ice-nucleating particles (DeLeon-Rodriguez et al., Reference DeLeon-Rodriguez, Lathem, Rodriguez-R, Barazesh, Anderson, Beyersdorf, Ziemba, Bergin, Nenes and Konstantinidis2013). Some studies also suggest that photosynthetic activity may be taking place in the upper parts of the troposphere, based on the presence of phototrophic organisms (Amato et al., Reference Amato, Besaury, Joly, Penaud, Deguillaume and Delort2019). These findings highlight the potential for airborne microbes to participate in planetary-scale processes on Earth and potentially other celestial bodies.
Aerobiome studies have demonstrated significant vertical stratification in microbial diversity across atmospheric layers. Taxonomic analyses using 16S rRNA sequencing have identified dominant bacterial phyla such as Proteobacteria and Actinobacteria in near-surface air samples, with hypoxia-tolerant and UV/oxidative stress-resistant taxa becoming increasingly prevalent at higher altitudes. Such results illustrate the need to investigate aerobiomes across different altitude gradients to understand their dynamics and adaptability (Gusareva et al., Reference Gusareva, Gaultier, Premkrishnan, Kee, Lim, Heinle, Purbojati, Nee, Lohar, Yanqing, Kharkov, Drautz-Moses, Stepanov and Schuster2020; Rodó et al., Reference Rodó, Pozdniakova, Borràs, Matsuki, Tanimoto, Armengol, Pey, Vila, Muñoz, Santamaria, Cañas, Morguí, Fontal and Curcoll2024; Zhen et al., Reference Zhen, Krumins, Fennell and Mainelis2018). Sampling airborne microbes at high altitudes presents unique technical challenges and is typically accomplished using a combination of ground-based towers, research aircraft, and balloon-borne devices. For altitudes up to around 1,500 meters, high-volume air samplers and filtration systems mounted on meteorological towers or tethered balloons are commonly used, enabling collection at defined heights within the boundary layer and lower free troposphere (Drautz-Moses et al., Reference Drautz-Moses, Luhung, Gusareva, Kee, Gaultier, Premkrishnan, Lee, Leong, Park, Yap, Heinle, Lau, Purbojati, Lim, Lim, Kutmutia, Aung, Oliveira, Ng, Dacanay, Ang, Spence, Phung, Wong, Kennedy, Kalsi, Sasi, Chandrasekaran, Uchida, Junqueira, Kim, Hankers, Feuerle, Corsmeier and Schuster2022; Spring et al., Reference Spring, Docherty, Domingue, Kerber, Mooney and Lemmer2018). At higher elevations – including the free troposphere transition layer (1,500–5,000 m) and especially the low stratosphere (∼12,000 m) – specialized aircraft equipped with bioaerosol collectors or remotely operated balloon-borne passive samplers are deployed to capture microbial biomass under extreme low-pressure and low-temperature conditions while minimizing contamination and ensuring altitude-specific sampling (Smith et al., Reference Smith, Ravichandar, Jain, Griffin, Yu, Tan, Thissen, Lusby, Nicoll, Shedler, Martinez, Osorio, Lechniak, Choi, Sabino, Iverson, Chan, Jaing and McGrath2018).
Sequencing techniques in aerobiome studies
High-throughput sequencing techniques have been widely applied to analyze airborne microbial communities across diverse environments, revealing seasonal dynamics and environmental influences on community composition. Advances in 16S rRNA sequencing techniques have revolutionized aerobiome research by enabling high-resolution taxonomic profiling of atmospheric microbial communities. It allows capturing subtle nucleotide variations in conserved ribosomal genes, improving strain-level differentiation while maintaining compatibility with universal primers that span conserved flanking regions (Johnson et al., Reference Johnson, Spakowicz, Hong, Petersen, Demkowicz, Chen, Leopold, Hanson, Agresta, Gerstein, Sodergren and Weinstock2019; Smith, Reference Smith2013; Woo and Yamamoto, Reference Woo and Yamamoto2020). For extreme environments like high-altitude aerobiomes, modified DNA extraction protocols and optimized primer sets now enable recovery of stress-tolerant taxa with reduced GC bias (Bharti and Grimm, Reference Bharti and Grimm2021; Rappaport and Oliverio, Reference Rappaport and Oliverio2023; Sierra et al., Reference Sierra, Ryon, Tierney, Foox, Bhattacharya, Afshin, Butler, Green, Thomas, Ramsdell, Bivens, McGrath, Mason and Tighe2022). These methodologies used on Earth directly inform astrobiological investigations. Portable 16S-targeted sequencing platforms and associated bioinformatics frameworks have been validated in Mars-analog permafrost environments, demonstrating the capacity to detect microbial activity through nucleic acid signatures under extreme cold and low humidity, which may pave the way for metagenomic studies in distant biomes in the future (Goordial et al., Reference Goordial, Altshuler, Hindson, Chan-Yam, Marcolefas and Whyte2017).
Understanding the metabolic activity of atmospheric microbes provides critical insights for developing biosignature detection frameworks applicable to planetary exploration. Terrestrial studies demonstrate that microbial redox reactions produce distinct gas byproducts like methane, nitrous oxide, and dimethyl sulfide. These exhibit temporal patterns correlated with metabolic processes and environmental conditions such as hypoxia and UV exposure (Chou et al., Reference Chou, Mahaffy, Trainer, Eigenbrode, Arevalo, Brinckerhoff, Getty, Grefenstette, Da Poian, Fricke, Kempes, Marlow, Sherwood Lollar, Graham and Johnson2021; Krissansen-Totton et al., Reference Krissansen-Totton, Olson and Catling2018; Olson et al., Reference Olson, Schwieterman, Reinhard, Ridgwell, Kane, Meadows and Lyons2018). Recent work by Dannenmann et al. with extremophile cultures has quantified threshold detection limits for metabolic intermediates under simulated exoplanetary conditions, revealing that instruments like impact ionization mass spectrometers can identify nucleobase patterns and lipid biomarkers at concentrations as low as 1 ppm in icy grain analogs (Dannenmann et al., Reference Dannenmann, Klenner, Bönigk, Pavlista, Napoleoni, Hillier, Khawaja, Olsson-Francis, Cable, Malaska, Abel and Postberg2023). However, distinguishing biological gas production from abiotic sources requires contextual analysis of planetary redox states and energy gradients (National Academies of Sciences, Engineering, and Medicine et al., 2018). By characterizing how Earth’s atmospheric microbes alter local gas equilibria through methanogenesis, iron oxidation, and other metabolisms, researchers establish comparative benchmarks for interpreting anomalous atmospheric chemistries detected on exoplanets or icy moons (Thompson et al., Reference Thompson, Krissansen-Totton, Wogan, Telus and Fortney2022). Earth-atmosphere interaction suggests promising possibilities for developing specialized tools to detect signs of active metabolism and preserved molecular fossils in extraterrestrial aerosols.
Here, we present an analysis based on 51 studies, comprising 3,584 samples obtained from various atmospheric layers, ranging from the planetary boundary layer to the lower stratosphere. Using aggregated 16S rRNA sequencing data, we performed taxonomic profiling to characterize the microbial diversity across altitudinal gradients. To infer the functional potential of these aerobiomes, we employed PICRUSt2, a bioinformatics tool that predicts metagenomic content by inserting amplicon sequence variants (ASVs) into a reference phylogenetic tree and estimating gene family abundances based on evolutionary relationships. PICRUSt2 predictions are weighted by the relative abundance of each ASV and adjusted for 16S rRNA gene copy number. This approach enables the estimation of pathway profiles derived from both dominant and phylogenetically diverse rare taxa. However, because predictions are based on inferred gene content rather than direct metagenomic sequencing, the resulting functional profiles are sensitive to both the taxonomic composition and the phylogenetic diversity of input sequences. Therefore, predicted metabolic pathways should be interpreted as composite representations of the community’s functional potential, reflecting contributions from dominant groups as well as ecologically distinct minority taxa. By integrating taxonomic and predicted functional profiles, this study provides insight into the metabolic capacity of atmospheric microbial communities and their broader relevance to biosphere functioning and astrobiological processes.
Methods
Data collection
This project includes data available in Sequencing Read Archive (SRA), European Nucleotide Archive (ENA), and DNA Data Bank of Japan. Projects were searched by the following keywords: “air microbiome,” “air metagenome,” “aerobiome,” “air samples,” “aerosol metagenome,” “bioaerosols,” “aerosol microbiome,” “atmospheric microbiome,” “airborne microbial communities,” “airborne microbial genomics,” “microbial composition of air.” The goal of this study was to preserve homogeneity, reproducibility of the data, and high public availability, therefore, only studies involving 16S rRNA gene sequencing were analyzed. Metadata was collected using RunSelector by National Center for Biotechnology Information (NCBI). Data related to sample collection, sequencing method, and altitude were derived either from metadata provided by the authors or descriptions in studies directly related to BioProject number. All projects included in the study are listed in Supplementary Table 1.
Data processing
Projects were downloaded using the SRA toolkit and converted into FASTQ files for further analysis. Sequencing files were forwarded for the quality control step with FastQC (0.12.1) and MultiQC (1.25.2) report generation. Trimmomatic was used to discard sequences shorter than 50 bp and discard any possible sequencing adapters remaining. For 16S rRNA taxonomy, classification by Qiime2 (2024.5) (Bolyen et al., Bolyen et al., Reference Bolyen, Rideout, Dillon, Bokulich, Abnet, Al-Ghalith, Alexander, Alm, Arumugam, Asnicar, Bai, Bisanz, Bittinger, Brejnrod, Brislawn, Brown, Callahan, Caraballo-Rodríguez, Chase, Cope, Da Silva, Diener, Dorrestein, Douglas, Durall, Duvallet, Edwardson, Ernst, Estaki, Fouquier, Gauglitz, Gibbons, Gibson, Gonzalez, Gorlick, Guo, Hillmann, Holmes, Holste, Huttenhower, Huttley, Janssen, Jarmusch, Jiang, Kaehler, Kang, Keefe, Keim, Kelley, Knights, Koester, Kosciolek, Kreps, Langille, Lee, Ley, Liu, Loftfield, Lozupone, Maher, Marotz, Martin, McDonald, McIver, Melnik, Metcalf, Morgan, Morton, Naimey, Navas-Molina, Nothias, Orchanian, Pearson, Peoples, Petras, Preuss, Pruesse, Rasmussen, Rivers, Robeson, Rosenthal, Segata, Shaffer, Shiffer, Sinha, Song, Spear, Swafford, Thompson, Torres, Trinh, Tripathi, Turnbaugh, Ul-Hasan, van der Hooft, Vargas, Vázquez-Baeza, Vogtmann, von Hippel, Walters, Wan, Wang, Warren, Weber, Williamson, Willis, Xu, Zaneveld, Zhang, Zhu, Knight and Caporaso2019) was used with the Silva132 database. Amplicon sequence variants (ASVs) were reconstructed using the dada2 plugin for Qiime2. Taxonomy results were exported to BIOM format. Representative sequences and taxonomy assignments were used to predict metabolic pathways using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2 (PICRUSt2) (Douglas et al., Reference Douglas, Maffei, Zaneveld, Yurgel, Brown, Taylor, Huttenhower and Langille2020). Analysis with PICRUSt2 was performed using EU server of Galaxy online platform (Afgan et al., Reference Afgan, Baker, Batut, van den Beek, Bouvier, Cech, Chilton, Clements, Coraor, Grüning, Guerler, Hillman-Jackson, Hiltemann, Jalili, Rasche, Soranzo, Goecks, Taylor, Nekrutenko and Blankenberg2018). Data analysis was performed using Pandas 2.2.3, NumPy 1.26.4, stats 0.1.2a, and SciPy 1.15. Visualizations were prepared using Plotly, Seaborn 0.13, and Matplotlib 3.5.1. All p-values obtained from statistical tests, including Mann–Whitney U and Kruskal-Wallis tests, were adjusted for multiple comparisons using the Benjamini/Hochberg False Discovery Rate (FDR) correction, with an adjusted p-value < 0.05 considered statistically significant. Samples were analyzed based on the categories presented in Table 1.
Table 1. Sample allocation scheme based on altitude
Results
Taxonomy composition of bacterial classes
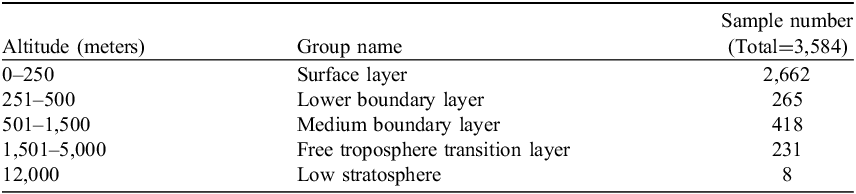
Bacterial class distribution varies markedly across atmospheric strata. In the Surface Layer, Alphaproteobacteria, Gammaproteobacteria, Actinobacteria, and Bacilli exhibit the highest relative abundances. The Lower Boundary Layer similarly contains elevated levels of Alphaproteobacteria and Gammaproteobacteria, while Actinobacteria and Bacilli show a moderate decline. In the Medium Boundary Layer, Gammaproteobacteria and Alphaproteobacteria remain prevalent, but most other classes – including Actinobacteria, Cyanobacteria, and Bacilli – show reduced abundance. The Free Troposphere Transition Layer is dominated by Alphaproteobacteria, with minor representation of Gammaproteobacteria and Actinobacteria. In the Lowest Stratosphere, Bacilli is the most abundant class, while most others, including Alphaproteobacteria and Actinobacteria, occur at low levels. Full taxonomy profiles are visualized in Figure 1; detailed data are available in Supplementary Table S1.
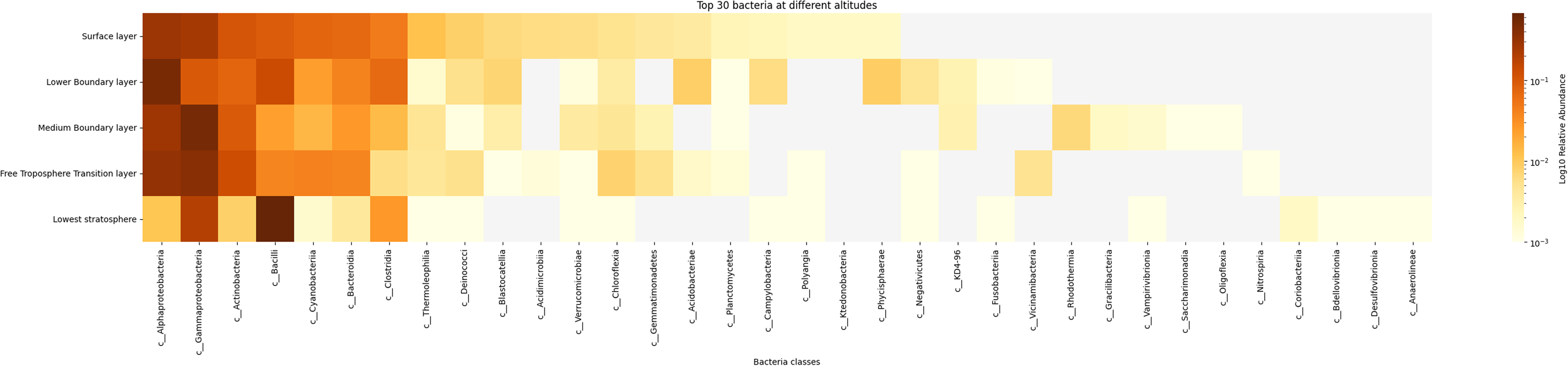
Figure 1. Relative abundances for the top bacterial classes across altitude layers (N = 3,584). Warmer colors indicate higher relative abundances, with distinct patterns observed for specific taxa at different altitudes. The Surface layer shows a broader distribution of bacterial classes compared to higher altitudes, reflecting variations in community composition along the vertical gradient.
Comparative analysis revealed statistically significant differences (low FDR) in microbial family abundances between the surface and the lower stratosphere. Several families exhibited marked enrichment in the stratosphere. Significant stratospheric enrichment was observed for Chromobacteriaceae (approx. 16.1-fold enrichment, FDR < 0.001), Coriobacteriaceae (approx. 7.8-fold enrichment, FDR < 0.001), Staphylococcaceae (approx. 42.6-fold enrichment, FDR < 0.001), and the archaeal family Methanobacteriaceae (approx. 4.0-fold enrichment, FDR < 0.001). The family Halomonadaceae, containing halophilic organisms, was also significantly more prevalent in the stratosphere, showing approximately 2.8-fold enrichment (FDR < 0.001). Conversely, families like Spirosomaceae (FDR < 0.001) were detected almost exclusively at the surface, indicating infinite enrichment at the surface relative to the stratosphere, although the statistical significance for these surface-dominant taxa was lower. Abundance of bacterial families is fully shown in Supplementary Table S2.
Alpha diversity of bacteria varies among altitudes
Bacterial diversity was determined using Shannon’s diversity index, which is an ecological metric derived from information theory that quantifies species diversity in a community by accounting for both the number of species (richness) and the relative abundance of each species (evenness). The Surface Layer exhibits the highest diversity, being represented by a wide range of values, while diversity decreases progressively with altitude. The Low Stratosphere shows the lowest Shannon diversity, reflecting a more uniform and less diverse microbial community. The boxplot (Figure 2) further illustrates these findings by depicting the distribution of diversity values across altitude layers.
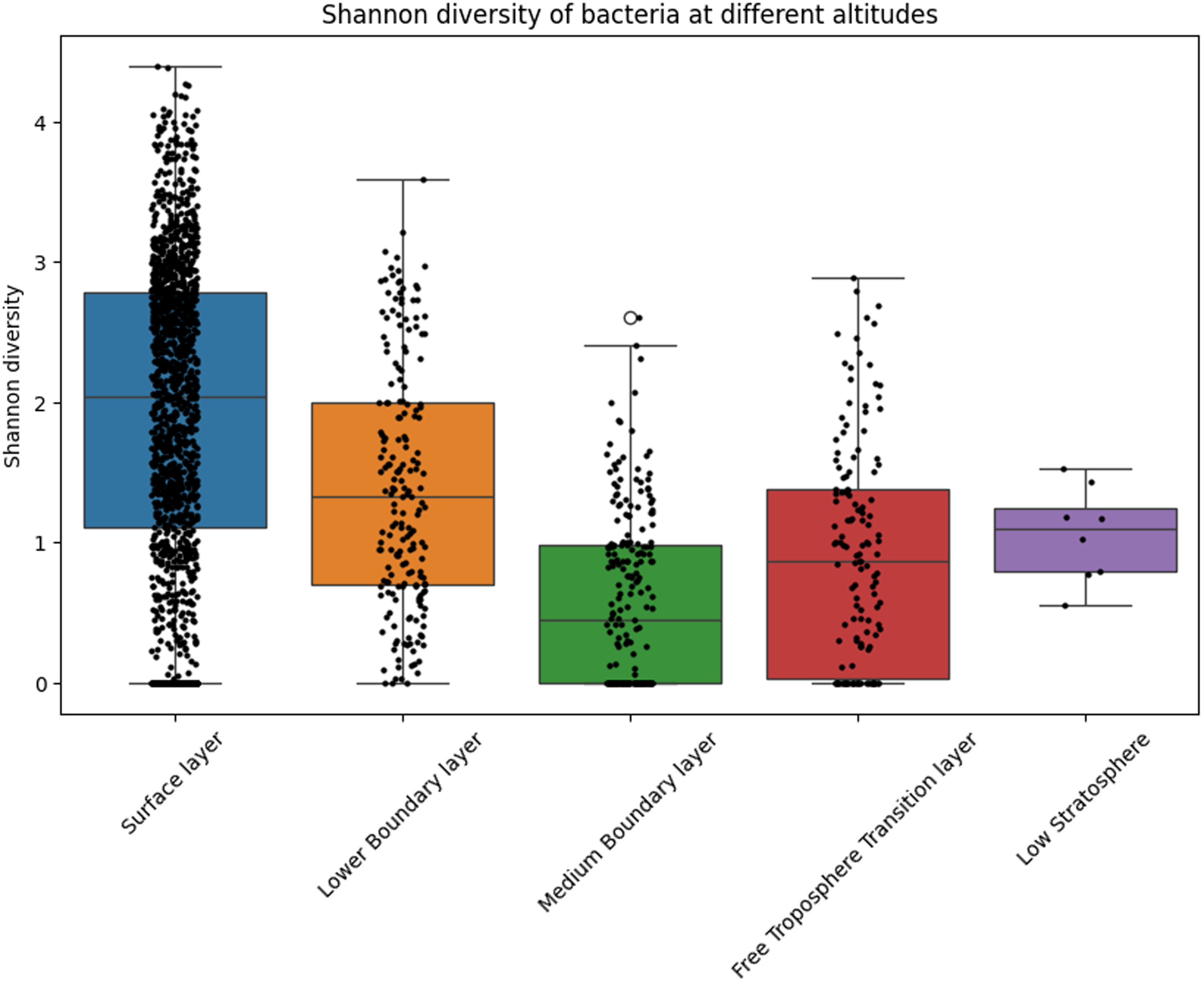
Figure 2. Shannon diversity of bacterial communities across altitude layers (N = 3,584). Each black dot represents the sample from the experiment. Dot with transparent background represents an outlier. The Surface layer displays the highest alpha diversity, with a broad range of Shannon diversity values, while diversity declines progressively with increasing altitude. The Low Stratosphere exhibits the lowest diversity, characterized by a narrower range and lower median values.
The Kruskal-Wallis test was applied to verify the Shannon index significance of bacterial communities across different altitude layers, revealing significant stratification in alpha diversity. The heatmap (Figure 3) presents the p-values for pairwise comparisons between altitude layers, with red indicating non-significant differences (FDR > 0.05) and green highlighting significant differences (FDR ≤ 0.05). Notably, no significant differences were observed between the Free Troposphere Transition Level and the Low Stratosphere (FDR > 0.05), suggesting similar microbial diversity in these upper atmospheric regions. In contrast, both layers exhibited significant differences when compared to all lower altitude layers, including the Surface layer, Medium Boundary layer, and Lower Boundary Level (FDR < 0.0001). Within the boundary layers, significant differences were also detected among the Surface, Medium Boundary, and Lower Boundary layers (FDR < 0.0001), highlighting distinct microbial communities at each level.
Pathway-specific analysis across investigated altitudes
In total, 467 individual metabolic pathways across 117 MetaCyc ontology pathway categories were predicted using PICRUSt2. To evaluate statistically significant differences in pathway activity across the atmospheric layers, the Mann–Whitney U test was applied to each pathway. All resulting p-values were adjusted for multiple comparisons using the Benjamini/Hochberg False Discovery Rate (FDR) correction method, with an adjusted p-value threshold of < 0.05 considered statistically significant. Analysis shows the number of statistically significant pathways (FDR < 0.05) shared between different altitude layers, with the highest overlap observed between the Medium Boundary Layer and Low Stratosphere (360 pathways). Surface and Lower Boundary Layers also share substantial similarity (328 pathways), while higher-altitude layers like the Free Troposphere Transition Layer and Low Stratosphere exhibit fewer overlaps with surface-associated layers. A full comparison is presented in Figure 4.
Figure 3. Kruskal-Wallis test p-values for Shannon diversity of bacterial communities across altitude layers (N = 3,584). The matrix compares pairwise differences in alpha diversity between the Free Troposphere Transition layer (N = 231), Low Stratosphere (N = 8), Lower Boundary layer(N = 265), Medium Boundary layer (N = 418), and Surface layer (N = 2,662).
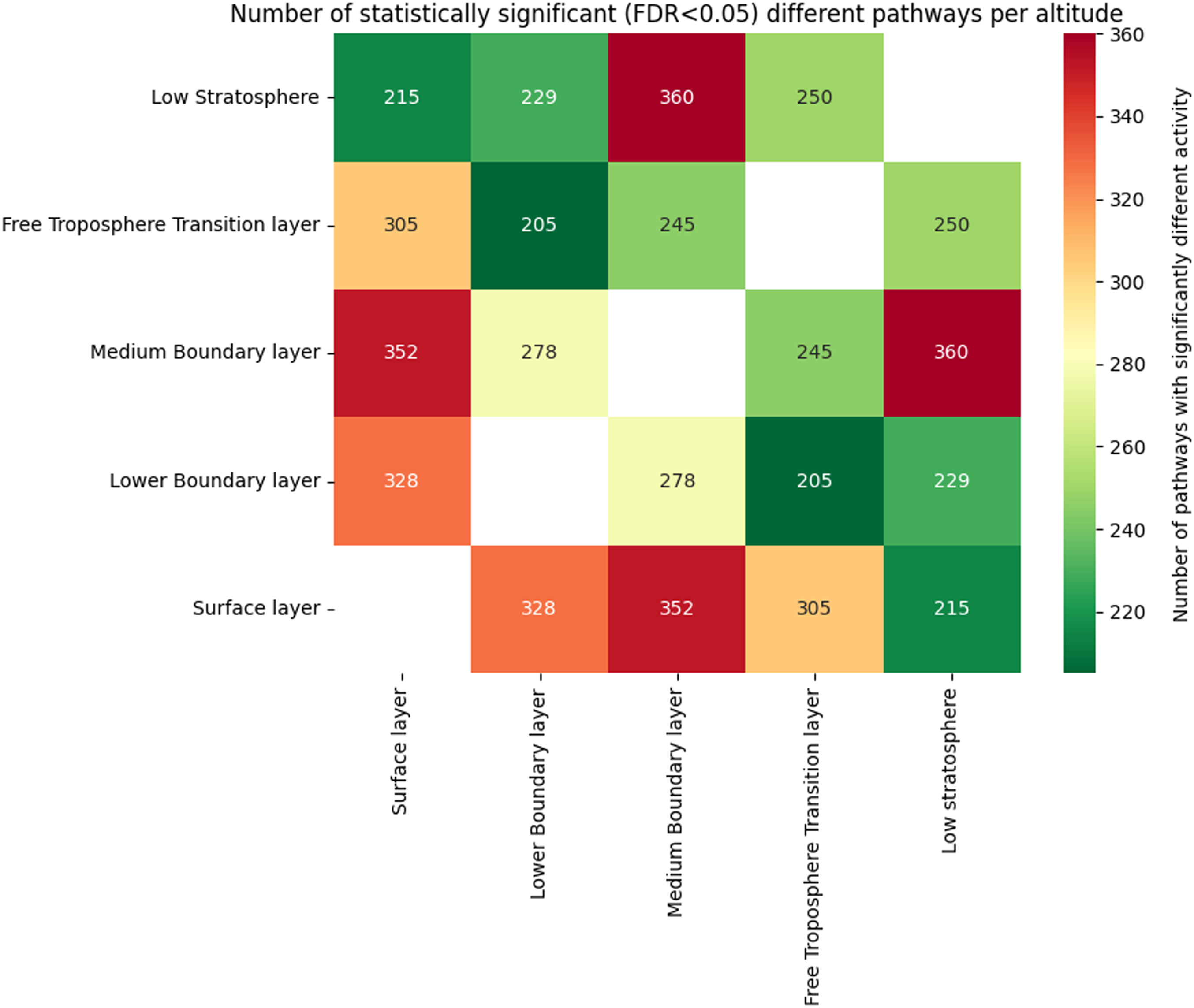
Figure 4. Number of statistically significant pathways (FDR < 0.05) with differential activity across various atmospheric altitude layers (N = 2,445). The color intensity corresponds to the number of significant pathways, with red indicating higher numbers and green indicating lower numbers.
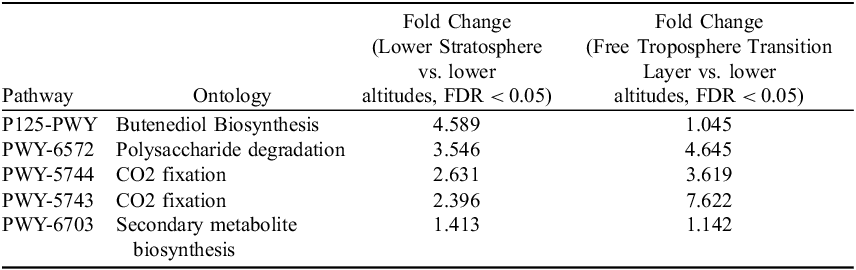
Microbial metabolic pathways in different atmospheric layers exhibit distinct activity patterns when compared to communities at lower altitudes, suggesting potential functional differences across the atmospheric gradient. In the Lower Stratosphere, Butenediol Biosynthesis (P125-PWY) exhibited the highest predicted activity (4.589-fold), alongside notable increases in Polysaccharide degradation (3.546-fold) and CO2 fixation via PWY-5744 (2.631-fold) and PWY-5743 (2.396-fold). These metabolic signatures could reflect adaptations for potential metabolic activity in the stratosphere, or alternatively, may represent genetic capacity for producing compounds that enhance cellular protection during dormancy or sporulation prior to atmospheric transport. In contrast, the Free Troposphere Transition Layer showed pronounced activity in CO2 fixation (PWY-5743: 7.622-fold) and Polysaccharide degradation (4.645-fold), indicating a stronger focus on carbohydrate metabolism and autotrophy in this region. Pathways like L-methionine biosynthesis (HSERMETANA-PWY: 1.148-fold) and Fatty acid degradation (FAO-PWY: 1.098-fold) were moderately upregulated in both layers, reflecting conserved roles in amino acid and lipid metabolism. Top 5 results are presented in Table 2, and all comparisons are available in Supplementary Table S3.
Table 2. Top 5 shared pathways between (Free Troposphere Transition Layer) and Lower Stratosphere in comparison to layers located at lower altitudes
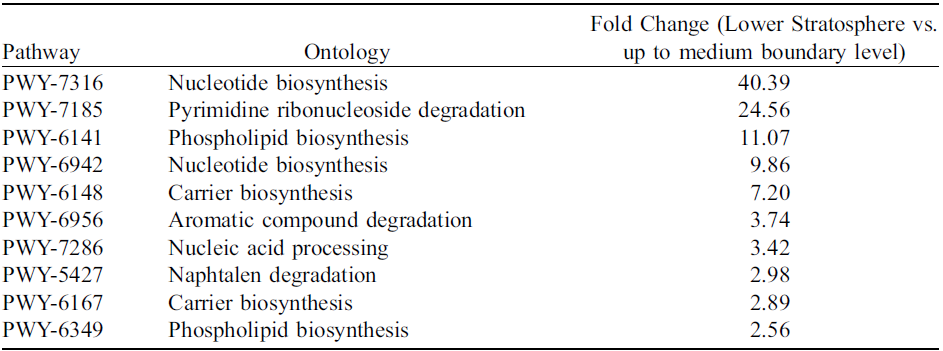
Microbial communities in the Lower Stratosphere demonstrated significantly elevated activity (FDR < 0.05) across multiple metabolic pathways compared to those at altitudes below 1,500 meters. The most pronounced differences occurred in nucleotide biosynthesis (PWY-7316: 40.39-fold; PWY-6942: 9.86-fold, FDR < 0.05) and pyrimidine ribonucleoside degradation (PWY-7185: 24.56-fold, FDR < 0.05). Pathways linked to phospholipid biosynthesis (PWY-6141: 11.07-fold; PWY-6349: 2.56-fold, FDR < 0.05) and carrier biosynthesis (PWY-6148: 7.20-fold; PWY-6167: 2.89-fold, FDR < 0.05) also showed marked upregulation. Additionally, increased activity was observed in aromatic compound degradation (PWY-6956: 3.74-fold, FDR < 0.05) and naphthalene degradation (PWY-5427: 2.98-fold, FDR < 0.05) alongside enhanced nucleic acid processing (PWY-7286: 3.42-fold, FDR < 0.05). These quantitative shifts highlight stratosphere-specific metabolic signatures distinct from lower atmospheric layers. Analysis of the top 10 mostly upregulated pathways is in Table 3.
Table 3. Shows metabolic pathways showing significantly increased activity (fold change, FDR < 0.05) in lower stratosphere samples compared up to medium boundary level
Conserved amino acid biosynthesis and lipid metabolism pathways define vertical metabolic stratification in low Earth atmospheric aerobiomes
The vertical stratification of microbial metabolic potential in low Earth’s atmosphere revealed distinct biosynthesis specializations in atmospheric layers (Figure 1). In the surface layer, L-isoleucine biosynthesis II (PWY-5101) exhibited the highest mean relative activity PICRUSt2 (8.3×10−3), followed by L-isoleucine biosynthesis I (ILEUSYN-PWY; 7.99×10−2) and L-valine biosynthesis (VALSYN-PWY; 7.99×10−2). The Lower Boundary Layer showed elevated activity in fatty acid elongation (FASYN-ELONG-PWY; 8.96×10−3) alongside persistent isoleucine biosynthesis pathways (PWY-5101: 9.78×10−3). A metabolic shift occurred in the medium boundary layer, where gondoate biosynthesis (PWY-7663; 1.099×10−2) and cobalamin salvage (PWY-5973; 9.73×10−3) dominated. The free troposphere transition layer has high activity in branched-chain amino acid synthesis (BRANCHED-CHAIN-AA-SYN-PWY; 8.13×10−3) and L-isoleucine biosynthesis I (9.62×10−3). The microorganisms in the lowest stratosphere retained peptidoglycan maturation (PWY0-1586; 7.2×10−3) while preserving residual amino acid biosynthesis capabilities (PWY-5101: 7.96×10−3) (Supplementary Figure S1).
The energy-related pathways identified across atmospheric layers of Earth demonstrate distinct stratification patterns, with aerobic respiration (PWY-3781, 1.36×10−2 surface; 1.62×10−2 free troposphere), Fermentation to alcohol (PWY-7111, 9.7×10−3 surface; 1.25×10−2 free troposphere), and pentose phosphate pathway (NONOXIPENT-PWY, 6.8×10-3 surface; 8.8×10−3 medium boundary layer) exhibiting the highest relative abundances in surface and transition layers. Core metabolic pathways such as the tricarboxylic acid (TCA) cycle (6×10-3–7.3×10−3), glycolysis (4.9×10−3–6×10−3), and glyoxylate bypass (3.5×10−3–4.7×10−3) maintain consistent activity profiles throughout the atmospheric column, while specialized pathways like fermentation (8x10−4–3.8×10−3) and denitrification (3×10−4–4×10−4) show reduced but measurable prevalence. Stratospheric Layer exhibits marked depletion in oxygen-dependent pathways (FERMENTATION-PWY: 3.7×10−3 lowest stratosphere vs 2.9×10−3 surface, FDR < 0.05), contrasting with elevated activity of transitional metabolic nodes (P161-PWY: 7.5×10−3 stratosphere vs 2.7×10−3 surface, FDR < 0.05) (Supplementary Figure S2). This vertical metabolic architecture demonstrates conserved amino acid biosynthesis machinery across strata, with lipid modification pathways becoming increasingly prominent at higher altitudes. Box plot presenting activity of ontology classes among altitudes is shown in Figure 5.
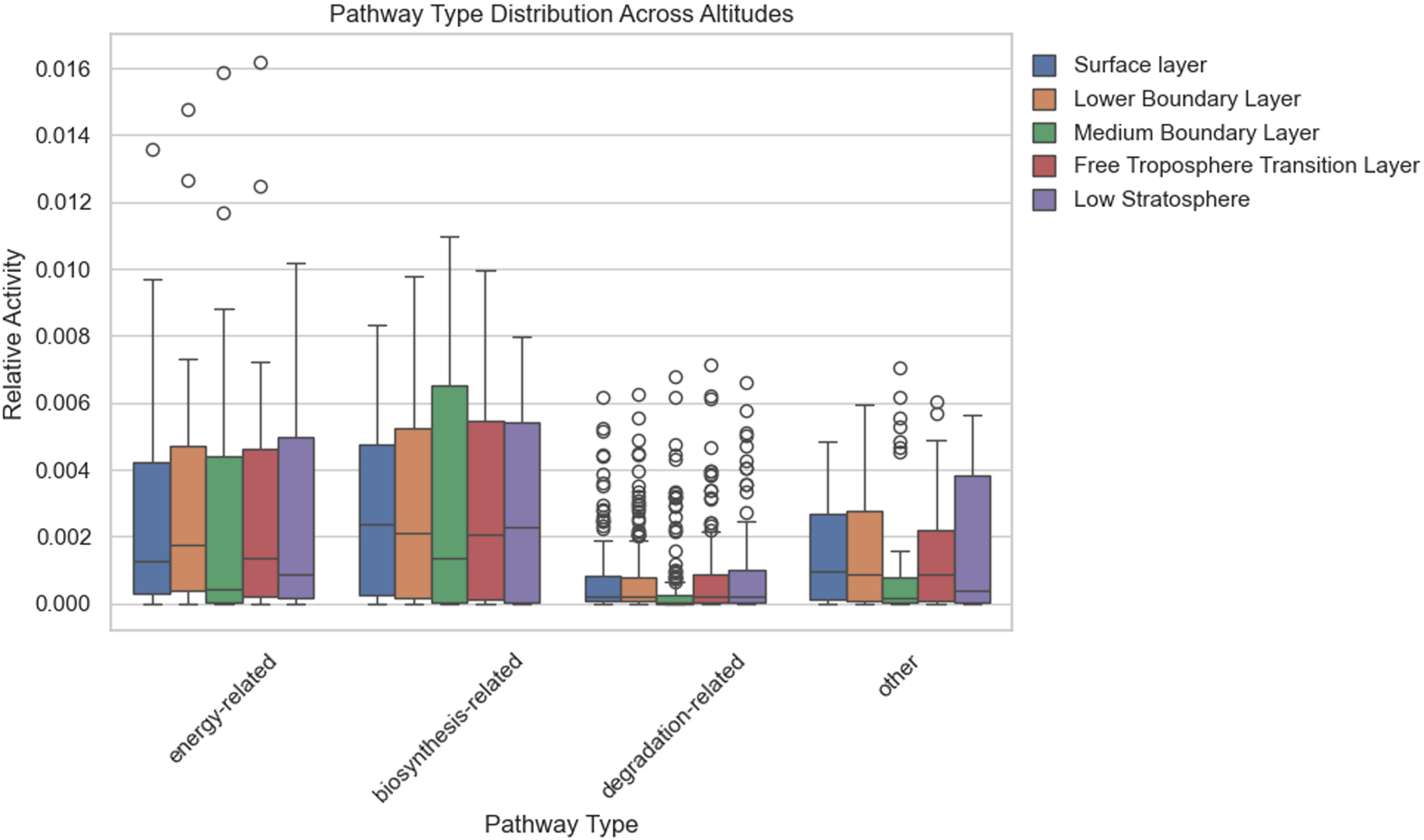
Figure 5. Relative activity of functional pathway types across five atmospheric altitude layers (N = 2,445). Each box represents the interquartile range, with whiskers indicating variability outside the upper and lower quartiles, and dots representing outliers.
Altitudinal variation in the relative presence of selected metabolic compounds
The predicted relative presence of specific compounds across altitudinal gradients, as inferred from pathway relative activity analysis, suggests potential patterns in their atmospheric distribution. Based on PICRUSt2 predictions, benzaldehyde showed a predicted relative presence ranging from 1.91×10-4 in the Surface Layer to a peak of 9.25×10−4 in the lowest stratosphere. Simultaneously, propionate showed a similar trend, increasing from 1.63×10−4 at the surface to a maximum of 8.10×10−4 in the stratosphere. Catechol concentrations varied between altitudes, with values spanning from 2.02×10−4 at the surface to a high of 7.69×10−4 in the stratosphere, suggesting differential environmental influences on its distribution. Ethanol (EtOH) displayed the most pronounced variation, rising from a surface value of 2.69×10−3 to a stratospheric maximum of 7.53×10−3, indicating significant altitude-driven changes in its presence. PICRUSt2 analysis suggests potential altitude-dependent variations in the predicted abundance of essential metabolites; the microbial communities at the lower boundary layer were predicted to have higher relative abundance of GTP (2.29×10−3), while communities sampled from the lowest stratosphere were predicted to have higher relative D-alanine pathway activity (5.03×10−3).
Vertical distribution of metabolic products across atmospheric layers
The predicted vertical distribution of key metabolic products across atmospheric layers suggests potential altitudinal patterns in microbial and biochemical activity, based on genomic inference rather than direct measurement. Isobutanol (Alcohols/Fermentation Products) exhibits peak relative presence (1.27×10−2) in the Lower Boundary Layer, consistent with enhanced fermentation processes under moderate oxygen availability. Fatty acid derivatives, such as 11Z-icos-11-enoyl-ACPs, show maximal abundance (1.1×10−2) in the Medium Boundary Layer, aligning with lipid metabolism requirements in intermediate atmospheric conditions. Fumarate (TCA Cycle Intermediates) maintains stable levels across layers (7.3×10−3–8.4×10−3) before declining in the stratosphere (6.3×10−3). Isoleucine (Amino Acids) demonstrates increased presence (8.5×10−3) in the Medium Boundary Layer, correlating with microbial redox homeostasis mechanisms under varying oxygen tensions. These patterns highlight metabolic trade-offs across altitudinal gradients, with fermentation products dominating lower atmospheric layers and lipid/amino acid derivatives showing mid-altitude maxima. The predicted lower abundance of certain metabolites in stratospheric samples (e.g., predicted isobutanol: 1.02×10−2 to 7.1×10−3) may reflect differences in microbial community composition under different atmospheric conditions. The data reveal altitude-specific optimization of microbial pathways, including TCA cycle modulation (fumarate) and branched-chain amino acid synthesis (isoleucine), likely driven by oxygen availability and temperature gradients. Complete data with estimated product abundance is available in Supplementary Table S4.
Discussion
Environmental conditions vary markedly across the atmospheric layers sampled in this study. The Surface Layer (0–250 m) and Lower Boundary Layer (251–500 m) are part of the atmospheric boundary layer, characterized by high air pressure, relative warmth, and substantial moisture and particulate content due to direct influence from the Earth’s surface and turbulent mixing processes. The Medium Boundary Layer (501–1,500 m) lies within the well-mixed portion of the boundary layer but begins to experience reduced surface influence, with decreasing temperature, humidity, and aerosol concentration. Above this, the Free Troposphere Transition Layer (1,501–5,000 m) marks the shift from the turbulent, well-mixed boundary layer to the more stable free troposphere, where air is cooler, drier, and less affected by surface emissions or mixing. The Low Stratosphere (∼12,000 m) is characterized by very low temperatures, extremely low humidity, much lower air pressure, and intense ultraviolet radiation, with minimal vertical mixing and a distinct increase in temperature with altitude due to ozone absorption of UV light. These gradients in temperature, pressure, humidity, and radiation create distinct ecological niches for airborne microbial communities at each altitude (De Wekker and Kossmann, Reference De Wekker and Kossmann2015; Kotthaus et al., Reference Kotthaus, Bravo-Aranda, Collaud Coen, Guerrero-Rascado, Costa, Cimini, O’Connor, Hervo, Alados-Arboledas, Jiménez-Portaz, Mona, Ruffieux, Illingworth and Haeffelin2023).
Vertical stratification of microbial diversity
The analysis of atmospheric microbial communities revealed pronounced altitudinal gradients in both taxonomic diversity and predicted metabolic potential. These observations suggest that Earth’s lower atmosphere contains distinct aerobiome communities with potentially different functional capacities, though the primary drivers of these differences remain to be fully elucidated. Bacterial alpha diversity, assessed via Shannon metrics, decreased significantly with altitude, peaking in the Surface Layer (highest richness/evenness) and reaching minima in the Low Stratosphere (FDR < 0.0001; Kruskal-Wallis test). Taxonomic composition shifted from Gammaproteobacteria, Alphaproteobacteria, Actinobacteria, and Bacilli dominance in the Surface and Lower Boundary Layers to Alphaproteobacteria, Gammaproteobacteria, and Bacilli prevalence in the upper layers, with Bacilli exhibiting the highest relative abundance in the Lowest Stratosphere. Metabolic pathway prediction analysis suggested potential differences between stratospheric and surface communities, including 4.6-fold higher predicted butenediol biosynthesis pathway activity and 2.6-fold higher predicted CO2 fixation pathway activity in stratospheric samples compared to surface layers. These findings align with prior studies documenting vertical microbial stratification in urban aerobiomes, but extend them by resolving metabolic mechanisms underlying high-altitude survival (Robinson et al., Reference Robinson, Cando-Dumancela, Antwis, Cameron, Liddicoat, Poudel, Weinstein and Breed2020, Reference Robinson, Cando-Dumancela, Liddicoat, Weinstein, Cameron and Breed2021). Additionally, statistical comparisons revealed that several microbial families – including uncultured Enterobacterales, Sulfurospirillaceae, Chromobacteriaceae, and Methanobacteriaceae – were significantly enriched in the stratosphere. The presence of these extremotolerant and metabolically versatile taxa in high-altitude aerobiomes underscores the potential for atmospheric microbial communities to serve as terrestrial analogs for life in extraterrestrial or otherwise extreme environments.
The decrease in observed diversity parallels studies linking reduced resource availability and harsher physicochemical conditions (e.g., UV radiation, desiccation) to bacterial community simplification at higher altitudes. However, an overlap of 360 metabolic pathways between the Medium Boundary Layer and Low Stratosphere suggests conserved metabolic strategies despite taxonomic divergence – a phenomenon potentially driven by atmospheric mixing or convergent evolution under shared stressors. Notably, the Free Troposphere Transition Layer and Low Stratosphere exhibited non-significant diversity differences (p > 0.05), contrasting with ecological boundaries reported in terrestrial systems (Hong et al., Reference Hong, Wu, Wilson and Song2018; Robinson et al., Reference Robinson, Cando-Dumancela, Antwis, Cameron, Liddicoat, Poudel, Weinstein and Breed2020). This homogenization may reflect microbial adaptation to extreme, stable conditions common to both layers. However, it is important to note that samples from the Low Stratosphere were obtained from only one project (Smith et al., Reference Smith, Ravichandar, Jain, Griffin, Yu, Tan, Thissen, Lusby, Nicoll, Shedler, Martinez, Osorio, Lechniak, Choi, Sabino, Iverson, Chan, Jaing and McGrath2018), with N = 8, which is a stark disproportion compared to the Free Troposphere Transition Layer, where N = 231.
The dominance of nucleotide biosynthesis (40-fold upregulation through PWY-7316 and 9-fold through PWY-6942) and phospholipid metabolism in the stratosphere implies microbial investment in genetic fidelity and membrane stability – critical for enduring low temperatures and oxidative stress. These adaptations mirror survival strategies observed in space-exposed extremophiles like Deinococcus radiodurans, which employ polyamine-mediated scavenging of reactive oxygen species and DNA repair mechanisms under extraterrestrial conditions (Tao et al., Reference Tao, Huang, Hungate, Manzoni, Frey, Schmidt, Reichstein, Carvalhais, Ciais, Jiang, Lehmann, Wang, Houlton, Ahrens, Mishra, Hugelius, Hocking, Lu, Shi, Viatkin, Vargas, Yigini, Omuto, Malik, Peralta, Cuevas-Corona, Di Paolo, Luotto, Liao, Liang, Saynes, Huang and Luo2023; Vincent et al., Reference Vincent, Fayolle, Hodyss, Johnson and Noell2024). The microbial communities detected in the stratosphere’s UV-rich, hypobaric environment display metabolic traits potentially relevant to astrobiological models of planetary habitability. These traits may result from selection pressures in the stratosphere, adaptive responses to source environments, or selective transport mechanisms. The observed metabolic resilience aligns with microbial survival in simulated Mars environments, where Pseudomonas aeruginosa and other species withstand desiccation and radiation and nucleotide salvage pathways (Zaccaria et al., Reference Zaccaria, de Jonge, Domínguez-Andrés, Netea, Beblo-Vranesevic and Rettberg2024).
Understanding of the bacterial metabolic pathways helps with biosignature investigation
The observed differences in predicted microbial metabolic pathways across Earth’s atmospheric layers may provide useful contexts for astrobiological research and biosignature interpretation. These metabolic profiles could reflect various selection pressures including, but not necessarily limited to, altitude-specific conditions. The distinct metabolic adaptations across atmospheric layers, such as enhanced CO2 fixation (PWY-5743) and secondary metabolite biosynthesis in the lower stratosphere, mirror strategies that extremophiles employ to thrive under resource-limited and oxygen-poor conditions. These findings align with studies suggesting that ancient metabolic pathways, like carbon fixation via reductive TCA cycles, may have been critical for early life on Earth and could similarly function in extraterrestrial settings (Noirungsee et al., Reference Noirungsee, Changkhong, Phinyo, Suwannajak, Tanakul and Inwongwan2024; Sokolskyi and DasSarma, Reference Sokolskyi and DasSarma2023). The predicted metabolic differences across atmospheric layers – such as higher representation of butanediol biosynthesis (P125-PWY, 4.589×) and CO2 fixation pathways (PWY-5743, 7.622×) in microbial communities from the lower stratosphere and free troposphere transition layer – may represent genetic capacity for energy and carbon metabolism under extreme conditions. These pathways could be relevant regardless of whether they are actively utilized in the atmosphere or primarily support pre-transport preparation and subsequent survival during atmospheric dormancy. For instance, butanediol can serve as both a fermentation product during active metabolism and a protective compatible solute during dormancy, while CO2 fixation capabilities could enable opportunistic carbon acquisition during periods of metabolic activity or contribute to cellular carbon reserves prior to dormancy. These findings align with studies on extremophiles, where genome-scale metabolic models (GEMs) have been used to predict metabolic responses to nutrient and energy limitations in extraterrestrial analog environments (Noirungsee et al., Reference Noirungsee, Changkhong, Phinyo, Suwannajak, Tanakul and Inwongwan2024). Increased abundance of secondary metabolites, such as aromatic compounds and naphthalene degradation products (PWY-6956: 3.74-fold, PWY-5427: 2.98-fold increase at higher altitudes), further underscores the biosynthetic versatility of aerobiomes. These pathways could serve as analogs for detecting complex organic molecules or biosignature gases on exoplanets (Abrahamsson and Kanik, Reference Abrahamsson and Kanik2022; Seager et al., Reference Seager, Schrenk and Bains2012). Additionally, the altitude-driven variations in compounds like ethanol and benzaldehyde reflect differential environmental influences on microbial activity, which could inform models predicting biosignature distributions under varying planetary conditions. Energy pathways such as glycolysis and the TCA cycle remain conserved across altitudes but exhibit reduced oxygen dependency at higher layers, mirroring metabolic shifts observed in anaerobic environments or prebiotic Earth analogs (Taha et al., Reference Taha, Patón, Penas, Banga and Rodríguez2023; Wimmer et al., Reference Wimmer, Xavier, Vieira, Pereira, Leidner, Sousa, Kleinermanns, Preiner and Martin2021).
Potential biosignatures and implications for extraterrestrial life detection
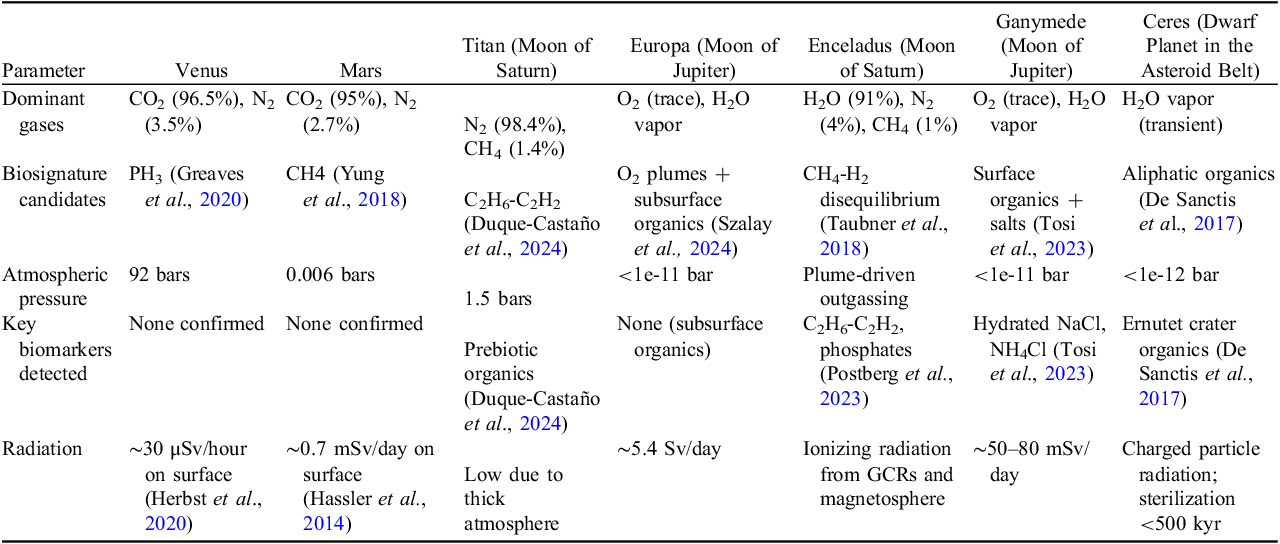
Earth’s atmospheric complexity – particularly the coexistence of O2 with reducing gases like CH4 – remains the gold standard for identifying biological disequilibrium – a state in which biological processes deviate from a stable condition due to environmental stress, evolutionary pressures, metabolic shifts, or disruptions in ecosystem dynamics. Results of this study reveal altitude-specific biosignature candidates with possible implications for extraterrestrial life detection. Higher altitude samples presenting significant upregulation of nucleotide biosynthesis pathways (PWY-7316, PWY-6942) and phospholipid biosynthesis (PWY-6141) suggest microbial adaptation to oxygen-poor conditions through conserved lipid modification strategies. These findings parallel methane-oxidizing metabolisms proposed for Enceladus’ subsurface ocean (Weber et al., Reference Weber, Marlin, Prakash, Teece, Dzurilla and Barge2023). Metabolic signatures, conserved across altitudinal gradients despite declining biomass, mirror the energy-efficient adaptations predicted for extremophiles in low-pressure, high-radiation environments. For example, the pronounced CO2 fixation in Earth’s Free Troposphere Transition Layer aligns with the proposed methanogenic or chemolithotrophic pathways in Mars’ subsurface brines, where CO2 and H2 disequilibrium could indicate microbial activity (Higgins and Cockell, Reference Higgins and Cockell2020; Westall et al., Reference Westall, Foucher, Bost, Bertrand, Loizeau, Vago, Kminek, Gaboyer, Campbell, Bréhéret, Gautret and Cockell2015). The predicted presence of TCA cycle intermediates (e.g., succinate) across Earth’s atmospheric layers, based on genomic inference rather than direct metabolite measurement, suggests potential alignment with proposals to consider glycolytic or fermentation byproducts as biosignatures in Martian brines or icy moon oceans, where oxygen-independent metabolism may dominate (National Academies of Sciences, Engineering, and Medicine et al., 2018; Wang and Qin, Reference Wang and Qin2024). Similarly, the predicted higher relative abundance of benzaldehyde and ethanol in stratospheric microbial communities may be considered in relation to the aliphatic organics detected on Titan, suggesting that gas-phase metabolic byproducts in icy moon plumes (e.g. Enceladus’ H2O/CH4/N2 jets) may serve as indirect biosignatures when coupled with energy-yielding pathways like sulfate reduction (Seeburger et al., Reference Seeburger, Higgins, Whiteford and Cockell2023; Weber et al., Reference Weber, Marlin, Prakash, Teece, Dzurilla and Barge2023) Metabolic modeling (GEMs) further refines these predictions: the conserved L-isoleucine biosynthesis (PWY-5101) and fatty acid elongation (FASYN-ELONG-PWY) observed across Earth’s atmospheric layers could guide targeted searches for amino acid isomers or lipid chains in Europa’s hypothesized subglacial redox gradients (Aguzzi et al., Aguzzi et al., Reference Aguzzi, Cuadros, Dartnell, Costa, Violino, Canfora, Danovaro, Robinson, Giovannelli, Flögel, Stefanni, Chatzievangelou, Marini, Picardi and Foing2024; Thakur et al., Reference Thakur, Singh and Zhang2022). What is more, the detection of hydrated NaCl and NH4Cl in Enceladus’ plumes with Earth-based halophile metabolisms reliant on osmolytes, underscores the utility of pathway-specific biomarkers (e.g., ectoine synthase activity) over generic organics (Baqué et al., Reference Baqué, Backhaus, Meeßen, Hanke, Böttger, Ramkissoon, Olsson-Francis, Baumgärtner, Billi, Cassaro, de la Torre Noetzel, Demets, Edwards, Ehrenfreund, Elsaesser, Foing, Foucher, Huwe, Joshi, Kozyrovska, Lasch, Lee, Leuko, Onofri, Ott, Pacelli, Rabbow, Rothschild, Schulze-Makuch, Selbmann, Serrano, Szewzyk, Verseux, Wagner, Westall, Zucconi and de Vera2022; Cleaves et al., Reference Cleaves, Hystad, Prabhu, Wong, Cody, Economon and Hazen2023). However, abiotic processes – such as photochemical synthesis of C2H6 on Titan or Martian perchlorate-driven CH4 oxidation – necessitate Bayesian frameworks to assess biosignature confidence (Schwieterman et al., Reference Schwieterman, Kiang, Parenteau, Harman, DasSarma, Fisher, Arney, Hartnett, Reinhard, Olson, Meadows, Cockell, Walker, Grenfell, Hegde, Rugheimer, Hu and Lyons2018; Vickers et al., Reference Vickers, Cowie, Dick, Gillen, Jeancolas, Rothschild and McMahon2023). Integrating in situ metabolomics with machine learning classifiers could resolve ambiguities, as demonstrated in this study by the Earth’s troposphere transition layer data showing co-upregulation of nucleotide biosynthesis and phospholipid pathways (PWY-6141), a pattern unlikely to arise abiotically. These approaches are critical for missions targeting Venusian cloud PH3 or Ceres’ Ernutet organics, where metabolic context (e.g., redox couples, ATP analogs) would distinguish living systems from prebiotic chemistry (Table 4) (Chandru et al., Reference Chandru, Potiszil and Jia2024; Hou et al., Reference Hou, Liu, Xu, Pang, Wang, Qin, Liu, Zhao, Wei, Xu, Jiang, Hao, Ji, Zhu, Yu, Liu, Sheng, Wang, Zhang and Li2024).
Table 4. The atmospheric compositions, biosignature candidates, pressures, and key biomarkers of potential celestial bodies that can become targets for further extraterrestrial life search missions
Metabolic capacity versus metabolic activity in atmospheric microbes
Our findings on taxonomic composition reveal the notable presence of spore-forming bacteria (Bacilli and Clostridia) in lower – stratospheric samples (Figure 1), consistent with the long-standing hypothesis that many high-altitude microbes exist primarily as dormant cells or spores. The predicted metabolic differences we observe could be interpreted through two complementary frameworks: (1) they represent the potential for active metabolism under stratospheric conditions, or (2) they reflect pre-transport physiological preparation that enhances survival during dormancy. For instance, the elevated nucleotide biosynthesis pathways (40-fold upregulation through PWY-7316 and 9-fold through PWY-6942) may serve dual purposes –supporting DNA repair during active growth in radiation-rich environments or, alternatively, establishing a robust nucleotide pool prior to sporulation that enhances long-term genetic stability during atmospheric transport. Similarly, the predicted upregulation of phospholipid metabolism could indicate either active membrane maintenance under stratospheric conditions or pre-dormancy membrane modifications that increase resistance to desiccation, UV radiation, and temperature fluctuations. While this study cannot resolve the ongoing debate about the metabolic state of atmospheric microbes, our findings on predicted metabolic capacity remain valuable regardless of whether these microbes are actively metabolizing in the stratosphere or utilizing these pathways primarily during ground-based growth phases to prepare for atmospheric dispersal.
Summary
Ultimately, Earth’s altitudinal biodiversity gradient may serve as a Rosetta Stone for interpreting exoplanetary atmospheric spectra. Planetary atmospheres are not homogenous, with distinct layers that may create unique environmental conditions. Our observations indicate distinct shifts in dominant microbial taxa and associated metabolic pathways at different altitudes, suggesting that environmental filtering drives community assembly patterns. These findings underscore the value of multi-altitude sampling strategies in astrobiological studies, combined with powerful bioinformatics methods, as single-layer analyses may overlook critical biosignature distribution patterns.
Conclusions
-
Microbial diversity decreases with altitude, with the Surface Layer exhibiting the highest Shannon diversity and the Low Stratosphere showing minimal diversity due to harsh environmental conditions.
-
Taxonomic composition shifts vertically, with Gammaproteobacteria, Alphaproteobacteria, Actinobacteria, and Bacilli dominating the lower atmospheric layers, while in the upper layers, Alphaproteobacteria, Gammaproteobacteria, and Bacilli remain prevalent, with Bacilli exhibiting the highest relative abundance in the Lowest Stratosphere.
-
Metabolic pathway analysis identifies significant differences in predicted pathway activities across altitudes, including higher relative activity of CO2 fixation and secondary metabolite biosynthesis pathways in the lower stratosphere. These differences may reflect various selective pressures, possibly including but not limited to altitude-specific conditions.
-
The Free Troposphere Transition Layer and Low Stratosphere communities share similar predicted metabolic profiles despite taxonomic differences, suggesting potential functional convergence that may be influenced by environmental conditions, selective transport, or source environment characteristics.
-
Altitude-driven variations in metabolic compounds such as ethanol and benzaldehyde suggest differential microbial activity influenced by environmental gradients.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1473550425100049
Data availability statement
All projects and samples considered in this study are presented in Supplementary Table S5. The project was conducted only using data accessible on public repositories.
Acknowledgments
The authors extend sincere gratitude to the members of the Polish Astrobiology Society for their invaluable support throughout this research. Special thanks are owed for their critical feedback during project workshops, access to specialized analytical tools, and collaborative discussions that enriched the study’s interdisciplinary approach. The Society’s commitment to fostering astrobiology research in Poland directly contributed to the methodological rigor and scope of this work.
Funding statement
The work was supported by funding from the Polish Astrobiology Society.
Competing interests
All authors declare no conflicts of interests.











