Introduction
Nudibranchs are globally distributed marine gastropod molluscs, renowned for their vivid colours and striking forms, and considered among the most beautiful animals in the oceans. Approximately, 3000 valid species of nudibranchs have been recorded, and this number continues to rise as new species are described (Wägele and Klussmann-Kolb, Reference Wägele and Klussmann-Kolb2005). Despite their diversity, our understanding of nudibranchs in Vietnam remains limited. Few studies have focused on the nudibranch community in Vietnam, with studies on this group rarely published. For example, Martynov and Korshunova (Reference Martynov, Korshunova, Britayev and Pavlov2012) documented approximately 120 nudibranch species from Vietnam, including the new species Janolus savinkini. Subsequently, Tuyen (Reference Tuyen2013) listed 55 nudibranch species from coral reefs in central Vietnam. Previous studies on nudibranchs in Vietnam have primarily relied on morphological examination method for species identification, often neglecting molecular methods. This limitation has made it challenging to identify species with similar appearances. Halgerda is a diverse nudibranch genus found in the Indo-Pacific (Tibiriçá et al., Reference Tibiriçá, Pola and Cervera2018). In a recent investigation, Donohoo et al. (Reference Donohoo, Villalobos, Hallas and Gosliner2023) described 14 new species of the genus. Nevertheless, information on the species composition of this genus in Vietnam remains sparse. To date, only H. tessellata, H. wasinensis and H. willeyi have been recorded in the country (Martynov and Korshunova, Reference Martynov, Korshunova, Britayev and Pavlov2012; Tuyen, Reference Tuyen2013). Continued research is essential to understand their diversity and distribution in Vietnamese waters.
DNA-based identification has gained prominence worldwide as an effective tool for identifying species (Hebert et al., Reference Hebert, Ratnasingham and deWaard2003). DNA-based methods offer unprecedented accuracy owing to their inherently high resolution. Moreover, using DNA barcoding is crucial for accurately estimating biodiversity, and it represents a pivotal technique in identifying nudibranchs. This approach has facilitated the detection of new records and the description of new species (Do et al., Reference Do, Jung, Kil and Kim2020; Pola et al., Reference Pola, Miguel-González and Paz-Sedano2023). Given its effectiveness in identifying nudibranchs, the present study employs DNA barcoding alongside morphological examination to provide the first record of Halgerda batangas collected from the Con Dao Islands, Vietnam.
Materials and methods
Nudibranch specimens were collected from two coral reefs at depths of 4 and 7 m in waters off the Con Dao Islands, Vietnam via scuba diving. Detailed information on the specimens is presented in Table 1. Collected specimens were preserved in 10% formaldehyde for morphological examination, which were prepared, along with species descriptions, following the guidelines of Carlson and Hoff (Reference Carlson and Hoff2000). The radula used for scanning electron microscope examination was prepared following Do et al. (Reference Do, Jung, Kil and Kim2020). The photographs of the radula were captured using the Hitachi TM4000Plus II with backscattered electron mode. Additionally, foot tissue samples were stored in 95% ethanol for DNA extraction. Total DNA was extracted from each sample using the G-spin™ Total DNA Extraction Mini Kit (iNtRON Biotechnology, South Korea). Polymerase chain reaction (PCR) analysis was performed using the primers LCO1490/HCO2198 (Folmer et al., Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994) for a partial sequence of cytochrome c subunit 1 gene (COI) and the primers 16Sar-L/16Sbr-H (Palumbi, Reference Palumbi, Hillis, Moritz and Mable1996) for a partial sequence of 16S ribosomal RNA gene (16S). The PCR reaction mixture consisted of 10 μl of 2X PCR Master Mix Solution (i-Taq) (iNtRON Biotechnology, South Korea), 1 μl of each primer (10 pmoles μl−1), 100 ng of DNA and distilled water to a final volume of 20 μl. The amplification conditions were as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 45 s, annealing temperature at 45°C (COI) and 48°C (16S) for 45 s, extension at 72°C for 1 min and final elongation at 72°C for 5 min. PCR products were verified via electrophoresis and sequences were obtained using the 3500xL Genetic Analyzer (Thermo Fisher Scientific, USA).
Table 1. Nudibranch specimens collected and examined in this study

Consensus sequences were generated from forward and reverse sequences using Geneious Prime 2023 (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheun, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Meintjes and Drummond2012). Sequences of congeneric species were retrieved from GenBank to estimate genetic distances (Table S1). Sequence distances were calculated using MEGA X software via the K2P method (Kumar et al., Reference Kumar, Stecher, Li, Knyaz and Tamura2018). Assemble Species by Automatic Partitioning (ASAP) was employed to delineate putative species (Puillandre et al., Reference Puillandre, Brouillet and Achaz2021).
For phylogenetic analysis, species without COI or 16S sequences were excluded. The optimal model for phylogenetic tree reconstruction was determined using PartitionFinder 2 (Lanfear et al., Reference Lanfear, Frandsen, Wright, Senfeld and Calcott2017). Phylogenetic trees were constructed using the Bayesian inference (BI) method in MrBayes ver. 3.2.7a (Ronquist et al., Reference Ronquist, Teslenko, Mark, Ayres, Darling, Hohna, Larget, Liu, Suchard and Huelsenbeck2012). The BI analysis consisted of two runs with four chains for 10 million generations, a sampling interval of 100 generations, and a burn-in of 25%.
Results
Morphological identification
Halgerda batangas Carlson and Hoff, Reference Carlson and Hoff2000
(Fig. 1)
Halgerda batangas Carlson and Hoff, Reference Carlson and Hoff2000: 157–159, figs. 6–11; Gosliner et al., Reference Gosliner, Valdés and Behrens2018: 111.
Halgerda malesso Debelius, Reference Debelius1996: 257 (Non H. malesso Carlson & Goff, 1993).
Type locality: Mactan Island, Cebu, Philippines.
Distribution: Western and Central Pacific (Gosliner et al., Reference Gosliner, Valdés and Behrens2018).
Material examined: Two Halgerda batangas specimens were collected from the Con Dao Islands, Vietnam, on 29 May and 02 September 2023 (COI GenBank numbers: PP331423 and PP331424; 16S GenBank numbers: PQ067230 and PQ067231).
Description: The two specimens measured 52 and 45 mm in length, respectively. The living animal (Figure 1) was translucent white and ovate. Its dorsal surface featured a network of fine, solid, red-orange lines. Both ridges and tubercles were present on the mantle. Smaller tubercles were located next to larger tubercles or they occurred along the ridges. The rounded orange-capped tubercles were surrounded by white bands. The size of the tubercles decreased toward the mantle margin. Rhinophores and gills were translucent white, with scattered small dark brown spots. The foot margin was delineated by an orange band. The gill had four primary branchial leaf rachises, with dark brown spots scattered across it.

Figure 1. Photograph of Halgerda batangas collected in the Con Dao Islands, Vietnam. (A) Dorsal view; (B) ventral view. Scale bars: 10 mm.
The radula (Figure 2) of the dissected 52-mm specimen (IMER-NU0001) had a formula of 65 × 53.0.53. The innermost lateral teeth were small with broad bases and shorter hooks compared to the mid-lateral teeth (Figure 2A). The size of the lateral teeth gradually increased toward the mid-lateral teeth. The mid-lateral teeth (Figure 2B) were hamate with long, pointed hooks. The outermost three to four teeth were reduced (Figure 2C).
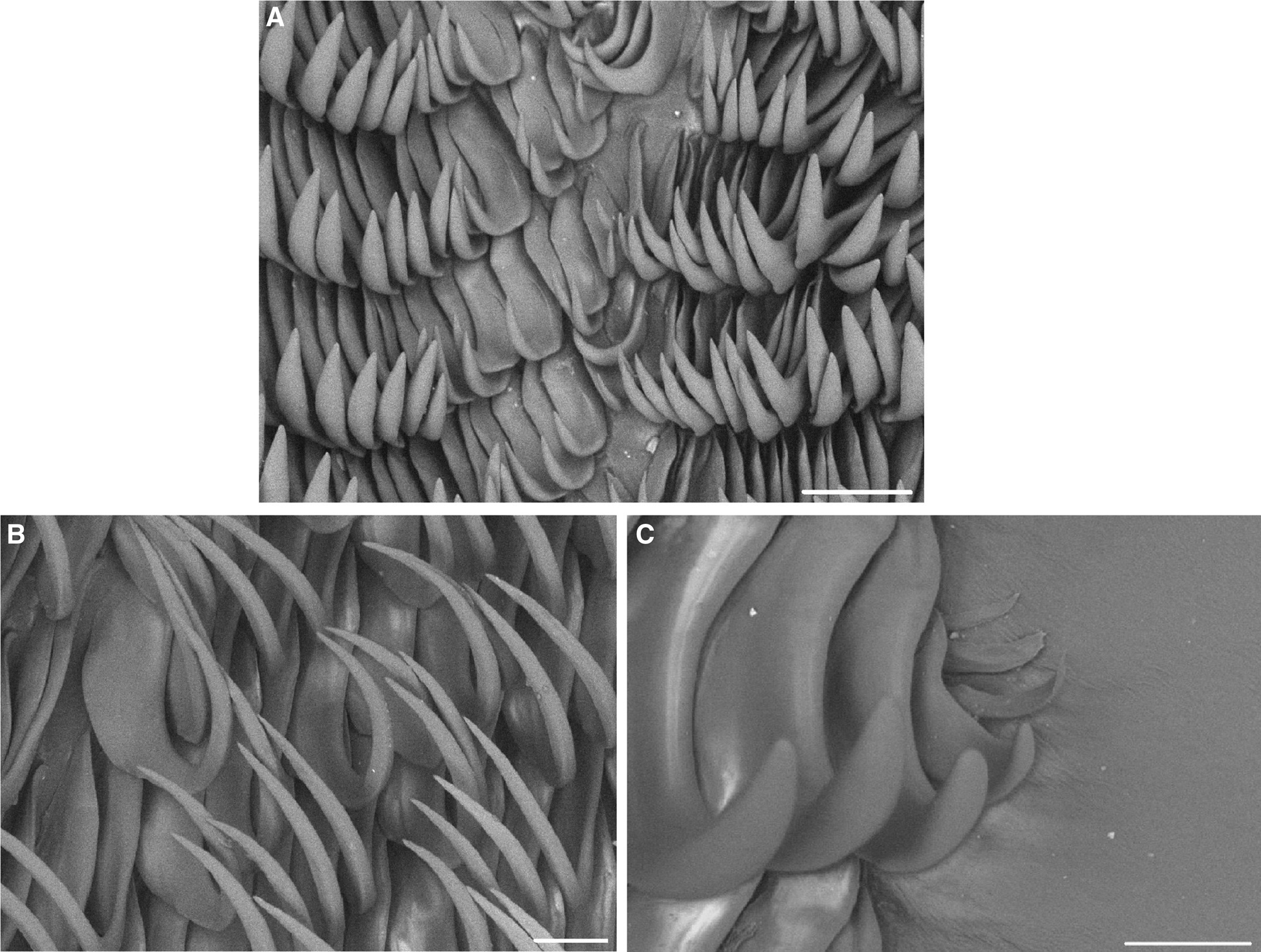
Figure 2. Radula of Halgerda batangas. (A) Inner lateral teeth; (B) mid-lateral teeth; (C) outer lateral teeth. Scale bars: 50 μm.
Remark
Two other species in the genus Halgerda, namely H. malesso and H. terramtuentis, are similar to H. batangas in having dorsal patterns of lines. H. batangas is characterized by relatively low tubercles with orange-red caps and white basal rings, along with an orange marginal band along the foot. In contrast, H. terramtuentis has a golden linear pattern, relatively low, white-capped tubercles on the dorsum, and a golden submarginal band along the mantle (Carlson and Hoff, Reference Carlson and Hoff2000). Conversely, H. malesso features orange lines on the dorsal surface, which may merge into orange markings in depressed areas of the dorsum, with relatively high tubercles (Carlson and Hoff, Reference Carlson and Hoff2000).
Analysis of COI and 16S sequences
The two COI sequences generated in this present study were 100% identical, resulting in 0% distance between the two sequences of Halgerda batangas. Intraspecific distances were 0–0.17%, whereas the minimum interspecific distance between H. batangas and other congeneric species was 1%, observed with H. malesso. ASAP analysis for species delimitation identified ten partitions, with H. batangas retrieved as a distinct taxonomic unit among the high number of subsets.
The 16S sequences showed 0% intraspecific distances, indicating that all examined 16S sequences were identical. However, the interspecific distances for 16S among H. batangas, H. malesso and H. diaphana were also 0%, indicating no difference in 16S across these species. ASAP analysis revealed 10 partitions, grouping H. batangas with H. malesso and H. diaphana owing to their sequence identities.
The phylogenetic tree for Halgerda constructed using BI (Figure 3) revealed that the specimens from this study formed a cluster with sequences obtained from GenBank. Specifically, H. batangas exhibited close relationships with congeneric species, including H. terramtuentis, H. malesso and H. diaphana. This suggests that the nudibranch samples collected in the present study belong to H. batangas.

Figure 3. Bayesian phylogenetic tree inferred from the partial COI and 16S gene sequences of Halgerda species. Accession numbers of COI sequences are placed in front of species names to indicate specific specimens shown in Table S1. Sequences generated in this study are marked with black spots. Carminodoris flammea and C. armata were used as outgroups. Posterior probability values are indicated at each node.
Discussion
Coral reefs are among the most biologically diverse marine ecosystems on Earth, hosting a wide range of reef organisms, including nudibranchs. However, nudibranchs from coral reefs in Vietnam remain under investigation. Most previous studies have compiled species lists, with only a few providing descriptions and illustrations. Continued research on nudibranchs is essential for understanding marine biodiversity in the country. In the present study, Halgerda batangas was recorded and described from the coral reefs of the Con Dao Islands, Vietnam. H. batangas belongs to a group in Halgerda known for its primarily white and orange markings. Originally described from the waters of the Philippines (Carlson and Hoff, Reference Carlson and Hoff2000), H. batangas from Vietnam exhibits characteristics consistent with those original descriptions, including a network of fine, solid, red-orange lines on the mantle (Carlson and Hoff, Reference Carlson and Hoff2000). It also features a white band around the mantle edge, with low, rounded, orange-capped tubercles. The mantle tubercles vary from small, rounded bumps to large, prominent structures. Previous studies in Vietnam have recorded three species of this genus: H. tessellata, H. wasinensis and H. willeyi (Martynov and Korshunova, Reference Martynov, Korshunova, Britayev and Pavlov2012; Tuyen, Reference Tuyen2013). Among these, H. tessellata is characterized by a yellow reticulated dorsum with small nodes (Martynov and Korshunova, Reference Martynov, Korshunova, Britayev and Pavlov2012; Tibiriçá et al., Reference Tibiriçá, Pola and Cervera2018) as well as white spots forming a band around the mantle (Tibiriçá et al., Reference Tibiriçá, Pola and Cervera2018). In contrast, H. wasinensis typically displays irregular dark blotches and a network of orange lines on the dorsum (Tibiriçá et al., Reference Tibiriçá, Pola and Cervera2018). Finally, H. willeyi has a complex dorsal pattern with yellow/orange ridging and black/dark brown radiating lines on the mantle (Tibiriçá et al., Reference Tibiriçá, Pola and Cervera2018; Donohoo et al., Reference Donohoo, Villalobos, Hallas and Gosliner2023). Meanwhile, H. batangas can be distinguished by its relatively low tubercles with orange-red caps and white basal rings, a reticulate network of orange lines on the dorsum, and an orange marginal band along the foot margin.
Consistent with the morphological findings, molecular analysis confirmed the presence of H. batangas in Vietnam. The genetic distance of COI sequences showed that intraspecific distances were lower than interspecific distances for the examined species. ASAP analyses also confirmed that H. batangas is a distinct species. However, 16S sequences could not differentiate H. batangas and its relatives owing to their high similarity. This finding indicates that 16S analysis alone is insufficient to distinguish H. batangas from closely related species. The phylogenetic tree, based on a combination of COI and 16S sequences, showed that all H. batangas samples were clustered together in a clade. Overall, the molecular phylogeny of Halgerda in the present study was congruent with the phylogenetic trees in the previous studies (Fahey, Reference Fahey2003; Tibiriçá et al., Reference Tibiriçá, Pola and Cervera2018; Donohoo et al., Reference Donohoo, Villalobos, Hallas and Gosliner2023). This result underscores the usefulness of molecular phylogeny for the genus. Therefore, expanding the sequence database for different species within the genus is necessary to investigate their phylogenetic relationships further.
Conclusion
This study confirms the new record of Halgerda batangas from the coral reefs of the Con Dao Islands, Vietnam, expanding the diversity of the nudibranch community in the country. Our results demonstrate the effectiveness of combining morphological and molecular approaches for investigating marine organisms. Given Vietnam's high marine diversity, this study highlights the potential for discovering new nudibranch species in the country's waters. Therefore, expanding surveys is crucial for a more comprehensive understanding of nudibranch diversity in Vietnam.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0025315424001176
Acknowledgments
We would like to thank Con Dao National Park staff for their support in the field survey.
Author contributions
T. D. D., T. C. D. and N. D. N.: conceptualization; H. M. T., M. V. N., L. M. N., J. R. P. and A. T. G.: field collection; T. D. D., H. M. T., M. V. N., J. R. P., A. T. G. and D.-w. J.: sample analysis and illustration preparation; T. D. D., Q. V. N. and T. C. D.: writing – original draft; T. D. D., L. L. W. and D.-w. J.: writing – review and editing. All authors approved the final version of the manuscript.
Financial support
The Vietnam Academy of Science and Technology (grant no. QTRU02.04/23-24) and the collaborative project between PharmaMar (Spain) and the Institute of Marine Environment and Resources (Vietnam) provided financial support for this study.
Competing interest
None.
Data
The nucleotide sequences generated during the current study were deposited in GenBank under accession numbers present in Table 1.






