Introduction
The family Echinostomatidae Looss, 1899 is composed of diverse digeneans that are globally distributed (Kostadinova, Reference Kostadinova, Jones, Bray and Gibson2005; Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019; Pantoja et al., Reference Pantoja, Faltýnková, O’Dwyer, Jouet, Skírnisson and Kudlai2021). Echinostomatids typically use molluscs as the first intermediate host, and the second intermediate hosts can be crustaceans, molluscs, amphibians, or fish, depending on the species (Tkach et al., 2016; Toledo & Esteban, Reference Toledo and Esteban2016). Adults of echinostomes have been reported from various vertebrates, with the highest diversity occurring in birds (Kostadinova & Jones, Reference Kostadinova, Jones, Jones, Bray and Gibson2005; Tkach et al., Reference Tkach, Kudlai and Kostadinova2016). Some echinostomatids belonging to Artyfechinostomum, Echinostoma, Echinoparyphium, Hypoderaeum, and Isthmiophora are intestinal parasites of humans who become infected by consuming raw or undercooked second intermediate hosts (Toledo & Fried, Reference Toledo, Fried and Motarjemi2014; Toledo & Esteban, Reference Toledo and Esteban2016). Echinostomatid infections in humans have been reported from several countries in Asia and Europe (Toledo & Fried, Reference Toledo, Fried and Motarjemi2014). Although species of Echinostoma, Echinoparyphium and Isthmiophora occur in Africa (Bisseru, Reference Bisseru1967; Appleton et al., Reference Appleton, Donnely and Eriksson1983; Toledo & Fried, Reference Toledo, Fried and Motarjemi2014; Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019), reports of human echinostomiasis are very few from the continent. Indeed, data on the infections in humans are available only from Kenya, Tanzania, and Egypt. According to Poland et al. (Reference Poland, Navin and Sarosi1985), a group of American tourists who had visited Kenya and Tanzania were diagnosed with echinostomiasis. However, the species that caused the infections were not identified (Poland et al., Reference Poland, Navin and Sarosi1985). In Egypt, human echinostomiasis is attributed to Echinostoma revolutum (Fröhlich, 1802) and Echinoparyphium recurvatum (von Linstow, 1873) (Toledo & Fried, Reference Toledo, Fried and Motarjemi2014). Considering the ecological importance and zoonotic potential of echinostomatids, they have been the subject of numerous investigations (Pinheiro et al., Reference Pinheiro, Maldonado Junior, Attias and Lanfredi2004; Toledo & Esteban, Reference Toledo and Esteban2016).
For many years, taxonomic knowledge of the family Echinostomatidae was based mainly on morphological characterisation of their adults (Kostadinova, Reference Kostadinova, Jones, Bray and Gibson2005; Kostadinova & Jones, Reference Kostadinova, Jones, Jones, Bray and Gibson2005). However, there has been considerable discussion on the morphological criteria used for species delimitation within Echinostomatidae, leading to revisions within the family (Pinheiro et al., Reference Pinheiro, Maldonado Junior, Attias and Lanfredi2004; Kostadinova, Reference Kostadinova, Jones, Bray and Gibson2005; Faltýnková et al., Reference Faltýnková, Gibson and Kostadinova2008a; Tkach et al., 2016). For instance, systematic relationships within the cosmopolitan genus Petasiger have been the subject of various studies. Although Faltýnková et al. (Reference Faltýnková, Gibson and Kostadinova2008a) recognised 18 Petasiger spp. following a comprehensive morphological study, phylogenetic analyses later inferred that Petasiger was polyphyletic. Thus, only 11 species: Petasiger azerbaydjanicus (Sailov, 1963); Petasiger carbonis (Mendheim, 1940); Petasiger exaeretus Dietz, 1909; Petasiger lobulatus Odhner, 1910; Petasiger mexicanus (Lamothe-Argumedo & Pérez-Ponce de León, 1989); Petasiger parvicephalus (Rietschel & Werding, 1978); Petasiger phalacrocoracis (Yamaguti, 1939); Petasiger radiatus (Dujardin, 1845); Petasiger segregatus (Dietz, 1909); Petasiger testitrifolius (Gogate, 1934); and Petasiger variospinosus (Odhner, 1910) were retained within the genus (Tkach et al., 2016). Unfortunately, molecular data are available only for three of the known species: P. exaeretus, P. phalacrocoracis, and P. radiatus (Tkach et al., 2016). In Africa, adult stages of Petasiger have been reported only for P. phalacrocoracis, P. variospinosus, and P. radiatus, from Tanzania, South Africa, and Zambia (Bisseru, Reference Bisseru1957; King & Van As, Reference King and Van As2000; Chibwana & Katandukila, Reference Chibwana and Katandukila2021). Because of the paucity of studies on adult specimens and absence of molecular data for most Petasiger spp., knowledge on the actual diversity and phylogenetic relationships within the genus remain incomplete (Tkach et al., 2016; Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019).
Similar to the adults, descriptions and identification of larvae of echinostomatids have largely been based on morphological characterisation. Unfortunately, identification of digeneans based on morphological descriptions of larvae alone often prove difficult or unreliable (Frandsen & Christensen, Reference Frandsen and Christensen1984; Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019). For instance, the taxonomic positions of many echinostomes from Africa remain uncertain because they were described using cercarial morphology and given provisional names without the assignment of generic names (Cawston, Reference Cawston1923; Faust, Reference Faust1926; Porter, Reference Porter1938; Fain, Reference Fain1953). In recent years, the incorporation of genetic data in studying intramolluscan stages of African echinostomes has proved beneficial for discriminating between morphotypes and providing information on their phylogenetic relationships (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019; Outa et al., Reference Outa, Sattmann, Köhsler, Walochnik and Jirsa2020; Schols et al., Reference Schols, Mudavanhu, Carolus, Hammoud, Muzarabani, Barson and Huyse2020; Hammoud et al., Reference Hammoud, Kayenbergh, Tumusiime, Verschuren, Albrecht, Huyse and Van Bocxlaer2022; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). On the other hand, comprehensive morphological data are lacking for most of those echinostomatids for which genetic data are available (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019; Schols et al., Reference Schols, Mudavanhu, Carolus, Hammoud, Muzarabani, Barson and Huyse2020; Hammoud et al., Reference Hammoud, Kayenbergh, Tumusiime, Verschuren, Albrecht, Huyse and Van Bocxlaer2022). Therefore, it is difficult to compare them with the species from earlier studies that were classified in the place holder genus ‘Cercaria’ (Cawston, Reference Cawston1923; Faust, Reference Faust1926; Porter, Reference Porter1938; Fain, Reference Fain1953).
Herein, echinostomatids are reported from Burnupia transvaalensis (Craven, Reference Craven1881), Burnupia trapezoidea (Boettger, 1910), and Burnupia mooiensis (Walker, Reference Walker1912) collected from the Vaal River (Orange River System) and Crocodile River (Limpopo River system), in South Africa. Morphological characterisation of the echinostomes was based on light and scanning electron microscopy (SEM). There is a paucity of data on the ultrastructural features of digenean parthenitae and cercariae (Pinheiro et al., Reference Pinheiro, Maldonado Junior, Attias and Lanfredi2004; Outa & Avenant-Oldewage, Reference Outa and Avenant-Oldewage2024). Therefore, in addition to optical data, the current study intended to assess the suitability of using tegumental features (observable only via SEM) for the differentiation of rediae and cercariae of closely related echinostomes. Taxonomic status of the echinostomes from this study were established using 28S rDNA sequences. The 28S rDNA gene possesses both variable and conserved regions and is useful for establishing boundaries between species and genera of diverse trematode families (Blasco-Costa et al., Reference Blasco-Costa, Cutmore, Miller and Nolan2016). Hence, the gene is the most widely used marker for inferring phylogenetic relationships between echinostomatids (Tkach et al., 2016; Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019; Izrailskaia et al., Reference Izrailskaia, Besprozvannykh and Tatonova2021). Additional genetic characterisations of the specimens were done using fragments of the ITS1-5.8S-ITS2 and 18S rDNA regions, and mitochondrial cytochrome c oxidase subunit 1 (cox1) gene. This follows the recommendation by Blasco-Costa et al. (Reference Blasco-Costa, Cutmore, Miller and Nolan2016) for a multi-loci characterisation of digeneans, to explore both interspecific and intraspecific variations, and provide a comprehensive reference database for future studies. Also, generation of new ITS and cox1 sequences allowed for the comparison of the echinostomatids from the present study with isolates from Zimbabwe (Schols et al. Reference Schols, Mudavanhu, Carolus, Hammoud, Muzarabani, Barson and Huyse2020; Mudavanhu et al., Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024), Kenya (Outa et al., Reference Outa, Sattmann, Köhsler, Walochnik and Jirsa2020), Tanzania (Chibwana & Katandukila, Reference Chibwana and Katandukila2021) and Uganda (Hammoud et al. Reference Hammoud, Kayenbergh, Tumusiime, Verschuren, Albrecht, Huyse and Van Bocxlaer2022), for which there are no 28S sequences.
Material and methods
Snail sampling and morphological analyses of digeneans
As shown in Fig. 1, the study was conducted at four sites, two each from the Vaal River (26.872364 °S, 28.117173 °E and 26.734854 °S, 27.634372 °E) and Crocodile River (25.959696 °S, 27.855555 °E and 25.957086 °S, 27.858308 °E), in South Africa. Snail sampling was done in summer (February and March) of 2022 and 2023 and in autumn (May 2023). Snails were picked by hand from submerged rocks and macrophyte stems, placed in plastic buckets containing pebbles and water from the sampling sites, and transferred to an onsite field laboratory. Identification of the snails was based on morphological features (Craven, Reference Craven1881; Walker, Reference Walker1912; Connolly, Reference Connolly1939; Brown, Reference Brown1994) and the cytochrome c oxidase subunit 1 mitochondrial gene (cox1). DNA sequences of the snails have been published elsewhere (Outa & Avenant-Oldewage, Reference Outa and Avenant-Oldewage2024). Isolation of digenean parthenitae and cercariae followed the procedures outlined by Frandsen and Christensen (Reference Frandsen and Christensen1984). Freshly isolated specimens were studied in temporary mounts; stained with Nile blue or unstained (Outa & Avenant-Oldewage, Reference Outa and Avenant-Oldewage2024). A drawing tube was used to make illustrations of each morphotype, followed by digitisation on Corel DRAW Graphics Suite X6 software (Corel Corporation, Ottawa, Canada). The specimens from the temporary mounts (representing different morphotypes from different snails) were transferred into 2-mL Eppendorf tubes containing 96% ethanol, for DNA analyses. Representative specimens of each morphotype from different snails (where possible) were preserved in 70% ethanol for morphometric analyses and SEM. Morphometric data of rediae and cercariae were obtained using a Zeiss Axioplan 2 epifluorescence microscope fitted with AxioVision 4.3 imaging software (Göttingen, Germany). Rediae and cercariae of each morphotype were prepared for SEM following the procedures provided by Nation (Reference Nation1983) and Outa and Avenant-Oldewage (Reference Outa and Avenant-Oldewage2024). The specimens were dehydrated in graded series of ethanol and hexamethyldisilazane (Merck, Darmstadt, Germany), mounted on adhesive conductive carbon tape fixed on glass microscope slides, and dried for 24 h in a Sanpla dry keeper desiccator cabinet (Kita-ku, Osaka, Japan). Gold coatings were applied on the mounted specimens using an Emscope SC500 (Quorum Technologies, Newhaven, UK) and a Vega 3 LMH, Tescan (Brno, Czech Republic) SEM was used to examine the specimens at 6 kV.

Figure 1. Map of the study area; adopted from Outa & Avenant-Oldewage (Reference Outa and Avenant-Oldewage2024). A, Southern Africa; B, Vaal River; C, Crocodile River. Site 1: below the Vaal Dam (26.872364 °S, 28.117173 °E); site 2: below the Vaal River Barrage Reservoir (26.734854 °S, 27.634372 °E); site 3: Lake Heritage (25.959696 °S, 27.855555 °E); and site 4: below Lake Heritage (25.957086 °S, 27.858308 °E).
Genetic and phylogenetic analyses
An E.Z.N.A. Tissue DNA Kit (Omega, Bio-tek, Inc, Georgia, USA) was used to extract genomic DNA based on the manufacturer’ s instructions. For each digenean morphotype, DNA was obtained from individual specimens of rediae and pooled samples of 10 cercariae per snail. Genetic characterisation was based on analyses of nuclear 18S, ITS and 28S rDNA, and cox1 gene. Polymerase chain reactions (PCRs) were performed in 30-μL volumes comprising 10 μL of DNA template, 3.8 μL of molecular grade water, 0.6 μL of each primer (forward and reverse), and 15 μL of Taq DNA Polymerase 2X Master Mix RED (Lasec) (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). Nuclear 28S rDNA, primers dig12 (5′-AAGCATATCACTAAGCGG-3′) and 1500R (5′-GCTATCCTGAGGGAAACTTCG-3′) (Tkach et al., Reference Tkach, Littlewood, Olson, Kinsella and Swiderski2003) were used, following the PCR conditions set by Outa et al. (Reference Outa, Bhika and Avenant-Oldewage2024). The Internal Transcribed Spacer (ITS) rDNA sequences consisting of ITS1-5.8S-ITS2 regions were amplified using BD1 (GTCGTAACAAGGTTTCCGTA) and BD2 (TATGCTTAARTTCAGCGGGT) (Luton et al., Reference Luton, Walker and Blair1992), in accordance with the PCR profile provided by Luo et al. (Reference Luo, Nie, Zhang, Wang and Yao2002). For 18S rDNA, amplification was done using primers JLR24 (5′-CGG AAT TCG CTA GAG GTG AAA TTC TTG G-3′) and JLR25 (5′-CCG AAT TCC GCA GGT TCA CCT ACG G-3′) (Campos et al., Reference Campos, Cummings, Reyes and Laclette1998). The PCR profile (Mwita & Nkwengulila, Reference Mwita and Nkwengulila2010) was modified by increasing the annealing temperature to 50 °C. Fragments of cox1 were amplified using primers Dice1F (5′-ATTAACCCTCACTAAATTWCNTTRGATCATAAG-3′) and Dice14R (5′-TAATACGACTCACTATACCHACMRTAAACATATGATG-3′) following the PCR profile described by Van Steenkiste et al. (Reference Van Steenkiste, Locke, Castelin, Marcogliese and Abbott2015).
Successful amplification of the PCR products was verified visually in 1% agarose gel, loaded with Safeview FireRed (Applied Biological Materials) dye. Gel electrophoreses were performed by applying 80V in a SmartDoc 2.0 ultraviolet trans illuminator (Benchmark Scientific, NJ, USA) for 30 minutes. Dye-terminator sequencing (Applied Biosystems, Warrington, Cheshire, UK) was done using forward and reverse primers and the products were purified in an ABI 3137 automated sequencer (Applied Biosystems) (Avenant-Oldewage et al., Reference Avenant-Oldewage, Le Roux, Mashego and Van Vuuren2014). The forward and reverse sequences were visually inspected, trimmed, aligned and assembled using Geneious Prime 2023.0.1, following the guidelines provided by Kearse et al. (Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Meintjes and Drummond2012). To identify isolates with the closest similarities to the sequences generated in the current study, nucleotide searches were conducted on the GenBank database using the Basic Local Alignment Search Tool (BLASTn). Sequences of echinostomatids on GenBank with at least 50% query cover were downloaded and aligned with sequences from the present study using MUSCLE program on the MEGA7 software. The alignments were trimmed and genetic divergence was compared in accordance with the procedures outlined by Tamura et al. (Reference Tamura, Stecher, Peterson, Filipski and Kumar2013). Lists of the sequences from GenBank that were compared with the current isolates are provided in Supplementary Tables S1-S4.
Phylogenetic trees were reconstructed using Bayesian inference (BI) and maximum likelihood (ML). The alignments that were used for phylogenetic analyses consisted of the sequences from the current study and representative sequences of echinostomatid genera, with species of the family Echinochasmidae as outgroup. In cases where multiple identical sequences of echinostomatids were available, only the sequences from adult worms (where present) or the longest sequences were included in the final alignments. Prior to the reconstructions, appropriate nucleotide substitution models were selected by running the final alignments through the model test tool in MEGA7. Accordingly, GTR+G (28S and ITS), JC+G+I (18S) and HKY+G (cox1) were applied. BI reconstructions were done in BEAST v2.5.0 (Bouckaert et al., Reference Bouckaert, Heled, Kühnert, Vaughan, Wu, Xie, Suchard, Rambaut and Drummond2014) by applying 10 million Markov chain Monte Carlo analysis. Convergence and effective sample size were checked using Tracer v1.7.1 (Rambaut et al., Reference Rambaut, Drummond, Baele and Suchard2018) and the Maximum Clade Credibility tree (50% posterior probability limit) inferred using TreeAnnotator v2.5.0. The ML phylograms were reconstructed in MEGA7. In all reconstructions, five categories of discrete gamma (G) distribution were applied, and the reliability of the nodal support was tested using 1000 bootstrap replicates.
Results
Of the 1645 specimens of Burnupia spp. that were examined, 1.22% were infected with echinostomes. Three echinostomatids (Petasiger radiatus [Dujardin, 1845], Petasiger sp., and Echinostomatidae gen. sp.) were identified (Table 1). There was no co-occurrence of different digenean species in individual snails. Morphological descriptions of the specimens are provided below. The current Petasiger sp. has been designated Petasiger sp. 3 ZA, to distinguish it from two other Petasiger spp. that were reported from lymnaeid snails from South Africa (Moema et al., Reference Moema, King and Baker2008; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024); these are herein designated Petasiger sp. 1 ZA and Petasiger sp. 2 ZA (Table 2). Morphometric comparisons between the rediae (n = 10) and cercariae (n = 20) of the current Petasiger spp., with specimens of Petasiger from other studies are provided in Tables 3 and 4. For Echinostomatidae gen. sp., cercariae were not observed; hence, descriptions are based on rediae (n = 7) that were isolated from a single snail. All measurements are presented in micrometres as means, followed by the minimum and maximum values in parentheses.
Table 1. Prevalence (%) of echinostomes in snails from the Vaal and Crocodile River systems

B. transvaalensis: S1, n = 590, S2, n =132; B. mooiensis: S2, n = 398; B. trapezoidea, S3, n = 128, S4 = 397; n.d., echinostomes not detected in the snails.
Table 2. List of cercariae of Petasiger spp. for which morphological descriptions are available, and their respective snail hosts and localities

Species from the current study are indicated in bold.
Table 3. Measurements (in μm) of rediae of Petasiger spp. from the current study (in bold) and previous studies, including species whose morphology correspond with Petasiger spp.

a Fain (Reference Fain1953).
b Lie & Basch, Reference Lie and Basch1967).
c Ostrowski de Núñez et al. (Reference de Núñez M, Hamann and Rumi1991).
d King and Van As (2000).
e Fernández et al. (2016).
f Outa et al. (Reference Outa, Bhika and Avenant-Oldewage2024).
g Našincová et al. (Reference Našincová, Scholz and Moravec1993).
Fixative/preservative: a, c and g, 4% formaldehyde solution; e, f and current specimens 70% ethanol; b and d, not stated.
Table 4. Measurements (in μm) of cercariae of Petasiger spp. from the current study (in bold) and previous studies, including species whose morphology corresponds with Petasiger spp.

a Fain (Reference Fain1953).
b Lie & Basch, Reference Lie and Basch1967).
c Ostrowski de Núñez et al. (Reference de Núñez M, Hamann and Rumi1991).
d King and Van As (2000).
e Moema et al. (Reference Moema, King and Baker2008).
f Fernández et al. (2016).
g Barton et al. (Reference Barton, Zhu, Nuhoglu, Pearce, McLellan and Shamsi2022).
h Outa et al. (Reference Outa, Bhika and Avenant-Oldewage2024).
i Našincová et al. (Reference Našincová, Scholz and Moravec1993).
Fixative/preservative: a, b and c – formaldehyde solution; f, g, h and current specimens 70% ethanol; d, not stated and e, live specimens.
Petasiger radiatus
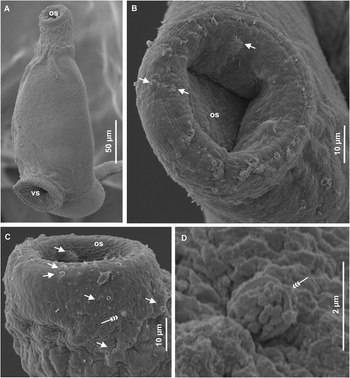
Redia whitish to brown, elongated, slightly curved dorsally, contain 6–9 cercariae (Fig. 2A). Mouth surrounded by five rows of papillae, bearing short sensilla (Fig. 3B). Region between anterior extremity and collar has numerous spherical bodies and sparsely distributed papillae with long sensilla (Fig. 3C). Pharynx, nearly spherical; digestive tube dark brown, extends posteriorly from pharynx, runs ventrally, 64% (60%–69%) of body length (Fig. 2A). Collar bears four (dorsoventral and two lateral) inconspicuous processes. Birth pore slightly protruded ( Fig. 3D), located on laterodorsal side of body, just posterior to collar. A pair of prominent ventral ambulatory buds (procruscula), located in posterior third of body.

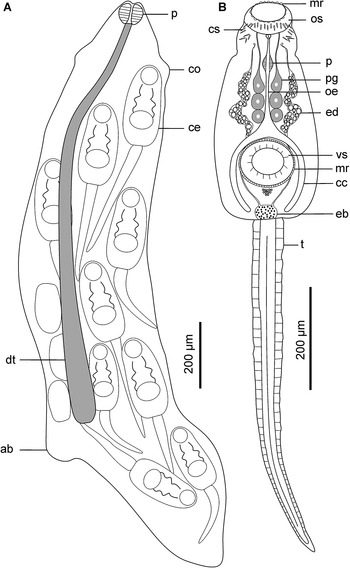
Figure 2. Schematic drawings of Petasiger radiatus. A, Redia and B, cercaria. Abbreviations: ab, ambulatory buds; cc, caecum; ce, cercaria; co, collar; cs, collar spines; dt, digestive tube; eb, excretory bladder; ed, main excretory duct; oe, oesophagus; os, oral sucker; mr, membranous rim; p, pharynx; pg, penetration gland cell; t, tail and vs, ventral sucker.

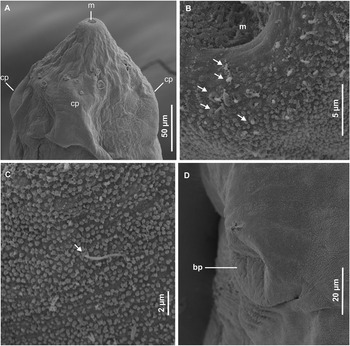
Figure 3. Scanning electron micrographs of redia of Petasiger radiatus. A, Lateral view of anterior end; B, rim of mouth; C, tegument structure on sub-apical end and D, birth pore. Arrows show uniciliated papillae. Abbreviations: bp, birth pore; cp, collar processes and m, mouth.
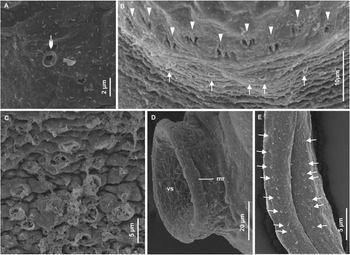
Cercarial body elongate-oval, widest near middle part (Fig. 2B). Collar bears 27 spines. General body surface aspinous. Oral sucker oval-shaped, surrounded by tegumental membranous rim (Fig. 2A and 4B). Eight to nine rows of uniciliated papillae present on tegument of anterior end: three on rim of oral sucker ( Fig. 4B), two on area between oral sucker and collar (Fig. 4C), 2–3 on collar and one posterior to collar (Fig. 5B). A pair of sub-apical multiciliated papillae (14–16 short cilia) present dorsolateral to oral sucker (Fig. 4D), cilia indistinct in some sensory receptors (Fig. 5A). Prepharynx characterised by a pair of prepharyngeal sacs immediately posterior to oral sucker. Pharynx ovoid; oesophagus long, bifurcates just anterior to ventral sucker; caeca terminate near posterior end of body (Fig. 2B). Six penetration gland cells present: three on each side of oesophagus. Ventral sucker post-equatorial, protrusible, transversely oval, larger than oral sucker, surrounded by tegumental membranous rim (Figs. 2B, 4A, and 5D). Genital primordium consists of an aggregation of cells posterior to ventral sucker. A pair of excretory ducts, each filled with 25–36 granules, extend anteriorly from excretory bladder (Fig. 2B). Tail simple, 1.7 (1.5–2.0) times longer than body; characterised by longitudinal furrow that extends from base and terminates near tip of tail. Tail tegument bears longitudinal rows of uniciliated papillae (Fig. 5C).

Figure 4. Scanning electron micrographs of cercaria of Petasiger radiatus. A, Ventral view of cercarial body; B, apical view of oral sucker; C, laterodorsal view of anterior end and D, close-up view of multiciliated papilla. Single arrows show uniciliated papillae and triple arrowheads show multiciliated papillae. Abbreviations: os, oral sucker and vs, ventral sucker.

Figure 5. Scanning electron micrographs of cercaria of Petasiger radiatus. A, Close-up view of papilla with a cluster of indistinct cilia; B, lateral view of collar, showing spines and papillae; C, ventral side of mid-region of the tail stem and D, sub-ventral view of the ventral sucker. Single arrows show uniciliated papillae, triple arrowheads show multiciliated papilla and arrow heads without tails show collar spines. Abbreviation: vs, ventral sucker.
Petasiger sp. 3 ZA
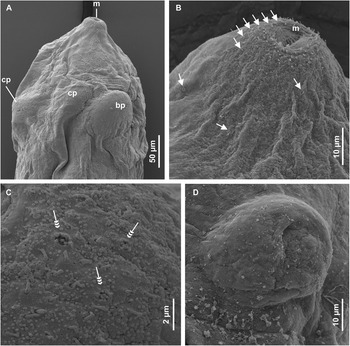
Redia whitish to orange, elongated, contain 5–8 developed cercariae (Fig. 6A). Mouth surrounded by 5–6 rows of sensilla (Fig. 7B). Each lateral side of mouth bears three multiciliated papillae, each consisting of 4–6 short cilia (Fig. 7C). Sparsely distributed papillae, bearing long sensilla occur between apical end and collar (Fig. 7B). Pharynx ovoid; digestive tube dark brown to black, extends ventrally from pharynx to 64% (61%–66%) of body length (Fig. 6). Collar bears four (dorsoventral and two lateral) short processes. Birth pore dorsal, prominently protruded, just posterior to collar (Fig. 7A and D). A pair of prominent ambulatory buds located ventrally, 67% (63%–73%) from anterior extremity.

Figure 6. Schematic drawings of Petasiger sp. 3 ZA. A, Redia and B, cercaria. Abbreviations: am, ambulatory buds; cc, caecum; ce, cercaria; co, collar; cs, collar spines; dt, digestive tube; eb, excretory bladder; ed, main excretory duct; mr, membranous rim; oe, oesophagus; os, oral sucker; p, pharynx; t, pg, penetration gland cell; tail and vs, ventral sucker.

Figure 7. Scanning electron micrographs of redia of Petasiger sp. 3 ZA. A, Dorsal view of anterior end; B, lateral view of apical end; C, papillae on lateral side of mouth and D, enface view of protrusion bearing birth pore. Single arrows show uniciliated papillae and triple arrow heads show multiciliated papillae. Abbreviations: bp, birth pore; cp, collar processes and m, mouth.
Cercarial body elongate-oval, widest near middle part; collar bears 27 spines (Fig. 6B). Entire body surface bears numerous minute spines, visible using SEM. Three rows of uniciliated papillae around oral sucker (Fig. 8B). A pair of sub-apical multiciliated papillae (18–22 cilia) present dorsolateral to oral sucker (Fig. 8B-E). Area between posterior margin of oral sucker and collar bears uniciliated papillae and unciliated pores (Fig. 8B-D). Lateral sides of body bear three rows of longitudinal uniciliated papillae. Oral sucker nearly spherical, surrounded by tegumental membranous rim (Figs. 6B and 8C). Prepharynx present, characterised by prepharyngeal sacs at posterior margin of oral sucker. Pharynx ovoid, oesophagus bifurcates into caeca at level of anterior margin of ventral sucker; each caecum terminates near posterior end of body (Fig. 6B). Penetration gland cells not clearly visible, appear to be five pairs along oesophagus. Ventral sucker post-equatorial, protrusible, transversely oval, larger than oral sucker (Figs. 6B, 8A, and 9D). Numerous cystogenous glands present, occurring from oesophageal region to posterior extremity. Secretions from glands visible (using SEM) on dorsal body surface on posterior part of some specimens (Fig. 9C). Two excretory ducts, filled with 30–42 granules, extend anteriorly from bladder towards pharyngeal region; flame cells pattern undiscernible. Tail, 1.5 (1.3–1.7) times longer than cercarial body, longitudinal furrow extends from tail base, terminates near tip. Tail tegument bears numerous minute spines and longitudinal dorso-ventral rows of uniciliated papillae (Fig. 9E).

Figure 8. Scanning electron micrographs of cercaria of Petasiger sp. 3 ZA. A, Lateroventral view of cercaria; B, dorsal view of anterior end; C, lateral view of anterior end; D and E, close up view of multiciliated papillae on dorsolateral side of anterior end. Single arrows show uniciliated papillae, winged arrowheads indicate unciliated pores and triple arrowheads show multiciliated papillae. Abbreviations: mr, membranous rim; os, oral sucker; t, tail and vs, ventral sucker.

Figure 9. Scanning electron micrographs of cercaria of Petasiger sp. 3 ZA. A, Close-up view of unciliated pore; B, dorsal view of collar; C, dorsal surface on posterior part of body; D, lateral view of ventral sucker and E, anterior part of tail stem. Single arrows show uniciliated papillae, winged arrowheads indicate unciliated pores and arrow heads without tails show collar spines. Abbreviations: mr, membranous rim and vs, ventral sucker.
Remarks on rediae and cercariae of Petasiger
Redial morphological characteristics of the two species described previously: sensilla around the mouth, collar with processes, birth pore posterior to collar, conspicuous ambulatory buds in the posterior third of the body, correspond with species of the family Echinostomatidae (Pinheiro et al., Reference Pinheiro, Maldonado Junior, Attias and Lanfredi2004; Keeler et al., Reference Keeler, Fried and Huffman2012; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). Cercarial morphological features: collar with 27 spines, two prepharyngeal granular sacs located on the posterior margin of the oral sucker, post-equatorial ventral sucker, suckers surrounded by tegumental membranous rim (crista), presence of granules in the main excretory ducts and a simple tail without finfolds, correspond with the genus Petasiger Dietz, 1909 (Našincová et al., Reference Našincová, Scholz and Moravec1993; Faltýnková et al., Reference Faltýnková, Nasincová and Kablásková2008b; Fernández et al., Reference Fernández, Hamann and Ostrowski-de Nunez2016; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024).
Redia of Petasiger sp. 3 ZA is distinguishable from Pet. radiatus by an ovoid pharynx and presence of multiciliated papillae around the oral aperture. The pharynx of Pet. radiatus is nearly round and multiciliated papillae were not observed. Cercaria of Petasiger sp. 3 ZA is distinguished by the presence of numerous tegumental spines on the body and tail and higher numbers of sensilla on the dorsolateral subapical papillae and penetration gland cells in the body, compared with Pet. radiatus. Morphological characteristics of the present Petasiger were compared with 16 cercarial morphotypes and redial data (where available) from 21 snail species in Africa, Europe, South America, and Australia (Table 2). This is inclusive of four echinostomatids whose cercarial features (collar with 27 spines, two prepharyngeal granular sacs on the posterior margin of the oral sucker, post-equatorial ventral sucker and presence of granules in the main excretory ducts) corresponds with Petasiger. They are: Cercaria bruynoghei and Cercaria decora (Fain, Reference Fain1953), Echinocercaria III (Ostrowski de Núñez et al., Reference de Núñez M, Hamann and Rumi1991) and Echinostomatidae sp. (Moema et al. (Reference Moema, King and Baker2008).
Rediae of different species are indistinguishable based on size due to overlap in body lengths between the species (Table 3). Rediae of the two species from the current study and Pet. variospinosus contain fewer cercariae compared with Petasiger sp. 2 ZA, C. bruynoghei and C. decora (Table 3). Differences were observed in the lengths of the intestinal tubes of some species. In Petasiger sp. from Argentina (Fernández et al. Reference Fernández, Hamann and Ostrowski-de Nunez2016) the intestinal tube extended only slightly into the posterior half of the body. For Pet. segregatus and the specimens in the current study, the intestine terminates in the posterior third of the body, just before the anterior margin of the ambulatory buds. In contrast, the intestinal tube extends to the level of the ambulatory buds in Pet. variospinosus (King & Van As, Reference King and Van As2000) and terminates posterior to the ambulatory buds in Petasiger sp. 2 ZA (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). Also, rediae of Petasiger sp. 2 ZA were characterised by a distinct papilliform process at the posterior extremity of the body (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024), while in the other species, the papilliform process was not apparent.
The length and width of cercarial body of Petasiger sp. 3 ZA were within the ranges of body dimensions of Pet. radiatus (current study) and four other species (Table 4). Only C. bruynoghei and Pet. segregatus were easily distinguished by their small sized bodies (Table 4). The morphology of Pet. radiatus cercaria from the current study is identical with the cercaria that was described by Našincová et al. (Reference Našincová, Scholz and Moravec1993) following a complete life cycle study of Pet. radiatus in the Czech Republic. However, the current cercariae are bigger (Table 4). Two cercarial morphotypes from Europe that were putatively identified as Paryphostomum radiatum syn. Pet. radiatus (Kiseliene, Reference Kiseliene1970; Faltýnková et al., Reference Faltýnková, Nasincová and Kablásková2008b) show considerable distinctions from the current specimens and the one described by Našincová et al. (Reference Našincová, Scholz and Moravec1993). The cercariae described by Kiseliene (Reference Kiseliene1970) and Faltýnková et al. (Reference Faltýnková, Nasincová and Kablásková2008b) were characterised by the presence of bifurcated excretory ducts in their tails. An excretory duct was not observed in the tail of the current cercaria nor in the specimens that were reported by Našincová et al. (Reference Našincová, Scholz and Moravec1993). The cercaria reported by Faltýnková et al. (Reference Faltýnková, Nasincová and Kablásková2008b) is further distinguished by at least 10 pairs of gland cells alongside the oesophagus. In contrast, the present specimens and those reported by Našincová et al. (Reference Našincová, Scholz and Moravec1993) were characterised by only three pairs of penetration gland cells. Also, contrary to the current cercaria in which sensory hairs were not observed using light microscopy and SEM, the species reported by Kiseliene (Reference Kiseliene1970) was characterised by tegumental sensilla that were visible using a light microscope. In this regard, the species described by Kiseliene (Reference Kiseliene1970) resembles cercariae of Pet. segregatus (Lie & Basch, Reference Lie and Basch1967) and Petasiger sp. (Fernández et al., Reference Fernández, Hamann and Ostrowski-de Nunez2016), both from South America, whose teguments are spinous.
The number of granules in each of the main excretory ducts of Petasiger sp. 3 ZA (30–42) is comparable with Pet. radiatus (25–36) (current study), Pet. radiatus (31–34) (Našincová et al., Reference Našincová, Scholz and Moravec1993), Petasiger sp. 2 ZA (27–38) (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024), and C. bruynoghei (35) (Fain, Reference Fain1953). These are distinct from other species which have fewer excretory granules, e.g. Petasiger sp. 2 (7–10), Petasiger sp. 4 (17) and Petasiger sp. 5 (19–20) (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019), Pet. variospinosus (19) (King & Van As, Reference King and Van As2000) and C. decora (21) (Fain, Reference Fain1953). Petasiger sp. 3 ZA is distinguished by five pairs of gland cells along the oesophagus. Fewer penetration gland cells were observed in Pet. radiatus (three pairs) and more in C. decora (13 pairs). Penetration gland cells were not discernible in Pet. segregatus, Echinocercaria III, Pet. variospinosus, Petasiger sp., Petasiger sp. 1 ZA and Petasiger sp. 2 ZA (Lie & Basch, Reference Lie and Basch1967; Ostrowski de Núñez et al., Reference de Núñez M, Hamann and Rumi1991; King & Van As, Reference King and Van As2000; Moema et al., Reference Moema, King and Baker2008; Fernández et al., Reference Fernández, Hamann and Ostrowski-de Nunez2016; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). The excretory systems of C. decora, Echinocercaria III, Pet. variospinosus and Petasiger sp. 1 ZA are characterised by 28 flame cells (Fain, Reference Fain1953; Ostrowski de Núñez et al., Reference de Núñez M, Hamann and Rumi1991; King & Van As, Reference King and Van As2000; Moema et al., Reference Moema, King and Baker2008). This is higher than in C. bruynoghei (24) and lower than in Pet. radiatus (30). Flame cell patterns were not clearly discernible in Pet. segregatus (Lie & Basch, Reference Lie and Basch1967), Petasiger sp. (Fernández et al., Reference Fernández, Hamann and Ostrowski-de Nunez2016), Petasiger sp. 2 ZA (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024), and Petasiger sp. 3 ZA (current study). Data for penetration gland cells and flame cells patterns are not available for the Petasiger spp. from Kenya (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019).
Apart from the current study, cercariae of only three other Petasiger spp. have been studied using SEM (King & Van As, Reference King and Van As2000; Moema et al., Reference Moema, King and Baker2008; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). Petasiger variospinosus is characterised by several short and long ciliated receptors surrounding the oral sucker, various groups of multiciliated papillae (3–23 short cilia) present dorsolateral to oral sucker and uniciliated papillae arranged bilaterally on both sides of the tail (King & Van As, Reference King and Van As2000). Petasiger sp. 1 ZA is distinguished by multiple clusters of 6–12 short cilia surrounding the oral sucker (Figure 3G, Moema et al., Reference Moema, King and Baker2008). Cercaria of Petasiger sp. 2 ZA is distinguished by two subapical papillae with few sensilla (up to four) and minute spines, scattered on the rest of the body (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). Petasiger sp. 3 ZA is characterised by numerous spines on the body and tail, and one pair of multiciliated papillae (18–22 cilia) on anterior end and numerous uniciliated papillae on the tail. Petasiger radiatus is distinguished by an aspinous tegument and a pair of anterior multiciliated papillae, each bearing 14–16 cilia.
Echinostomatidae gen. sp.
Redia orange, slightly curved dorsad (Fig. 10A and B), 1065 (915–1188) long, 241 (224–251) wide. Oral aperture surrounded by 6–7 rows of sensilla (Fig. 10C); lateral sides bear a pair of multiciliated papillae, each bearing 4–8 sensilla (Fig. 10D). Pharynx muscular, 52 (50–54) long, 47 (42–55) wide. Digestive tube dark brown to black, 511 (406–598) long, extends posteriorly, 53% (50%–55%) from anterior end. Collar, 149 (122–178) from the anterior end. Birth pore situated in pouch-like structure, on dorsal side just posterior to collar (Fig. 10B and E). Collar processes not observed; a pair of slightly protruded ambulatory buds located ventrally, 61% (58%–63%) from anterior end. Redia of this species is distinguished from Petasiger spp. by its short digestive tube (about half the body length) and a birth pore that is not elevated. What is more, the anterior end bears a pair of multiciliated papillae, contrary to three pairs in Petasiger sp. 3 ZA and Pet. radiatus in which multiciliated papillae were not observed.

Figure 10. Redia of Echinostomatidae gen. sp. A, Schematic drawing of whole body; B, scanning electron micrograph of whole body; C, apical view of anterior end; D, close-up view of oral papillae and E, enface view of birth pore. Single arrows show uniciliated papillae, triple arrowheads show multiciliated papillae. Abbreviations: am, ambulatory buds; bp, birth pore; ce, cercaria; co, collar; dt, digestive tube; m, mouth and p, pharynx.
Molecular and phylogenetic data
Usable rDNA sequences were obtained from seven, nine, and four isolates of Pet. radiatus, Petasiger sp. 3 ZA, and Echinostomatidae gen. sp., respectively. The newly generated sequences were 1214–1253, 1017–1029, and 871–898 bp for 28S, ITS, and 18S rDNA, respectively. The sequences have been submitted to GenBank: accession numbers PP738959-PP738964 (28S), PP738869-PP738871 (ITS), and PP738680- PP738682 (18S).
The 28S rDNA intraspecific variations for the sequences generated in the current study did not exceed 1 bp, corresponding to a p-distance of 0.1%. The 28S base pair differences and corresponding p-distances between the current sequences and other echinostomatids are shown in Supplementary Table S1. The p-distances between the current isolates of Pet. radiatus and sequences of Pet. radiatus obtained from adult worms (Tkach et al., 2016; Cech et al., Reference Cech, Molnár and Székely2017), ranged between 0% and 0.4%. The low variation between Pet. radiatus haplotypes was comparable to intraspecific variations between Pet. exaeretus isolates (0%–0.3%) published by Tkach et al. (2016) and Cech et al. (Reference Cech, Molnár and Székely2017). Petasiger sp. 3 ZA sequences differed from Pet. radiatus by p-distances of 1.2%–1.3%. Petasiger sp. 3 ZA showed the highest similarity (99.3%–99.4%) with cercaria of Petasiger sp. 5 from Bulinus globosus (Morelet, 1866) from Kenya (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019). Petasiger sp. 2 ZA (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024) varied from Petasiger sp. 3 ZA and Pet. radiatus by p-distances of 1.4–1.5 and 0.6–0.7 %, respectively. Echinostomatidae gen. sp. (current study) varied from other echinostomatids by p-distance ranges of 2.3%–5.7% (Supplementary Table S1). The 28S phylograms, consisting of 54 sequences of echinostomatids (1159–1170 bp), demonstrated that sequences of Petasiger occurred in 10 subclades (A–J) (Figs. 11 and 12). Petasiger spp. from South Africa clustered in three separate subclades (A, C, and D). Petasiger radiatus sequences (subclade A) were monophyletic with cercarial isolates of Petasiger sp. 4 from Kenya and Petasiger sp. from Australia (subclade B). However, the branching between clades A and B was poorly supported (0.45) in the BI tree (Fig. 11). Petasiger sp. 2 ZA clustered with Petasiger sp. 3 from Kenya and Petasiger sp. from Hungary in subclade C. Petasiger sp. 3 ZA-Petasiger sp. 5 clade was basal to A, B, and C. In both the BI and ML phylograms, cercarial isolate of Petasiger sp. 1 from Kenya formed a strongly supported subclade (J) with Pegosomum sequences that were obtained from adult worms. Petasiger exaeretus sequences were sister to the subclade comprising Pegosomum and Petasiger sp. 1. The positions of Petasiger sp. 2 from Kenya and sequences of Isthmiophora did not resolve clearly between the ML and BI frameworks. In the BI tree, Petasiger sp. 2 formed a poorly supported branch that was basal to the clade comprising of Pet. phalacrocoracis and Petasiger sp. 6. Also, Isthmiophora was sister to the Petasiger-Pegosomum clade (Fig. 11). In ML, Petasiger sp. 2 was sister to sequences of Isthmiophora in a weakly supported subclade (H), which was nested within the larger Petasiger clade (Fig. 12). Four species from Germany (KM191799- KM191807) whose cercariae were 19-spined and large tailed and were initially thought to belong to Petasiger (Selbach et al., Reference Selbach, Soldaánovaá, Georgieva, Kostadinova, Kalbe and Sures2014), clustered with Neopetasiger sequences. Echinostomatidae gen. sp. was positioned in a poorly supported clade containing sequences of Drepanocephalus, Chaunocephalus and Neopetasiger (Figs. 11 & 12).

Figure 11. Bayesian inference 28S rDNA phylogram of Echinostomatidae spp. The clades containing Petasiger spp. and Echinostomatidae gen. sp. are highlighted, and isolates from South Africa are indicated in bold. Nodal support values lower than 0.5 are not shown. Isolates marked with asterisks (**) are for 19-spined and large-tailed cercariae belonging to the genus Neopetasiger.

Figure 12. Phylogenetic relationships of Echinostomatidae spp. from the current study and from GenBank based on 28S rDNA inferred from maximum likelihood analyses. The clades containing Petasiger and Echinostomatidae gen. sp. are highlighted and isolates from South Africa are indicated in bold. Nodal support values lower than 50% are excluded. Isolates marked with asterisks (**) are for 19-spined and large-tailed cercariae belonging to the genus Neopetasiger.
The ITS rDNA sequences for each species were identical. Sequence divergence (%) and nucleotide substitutions between the current isolates and echinostomatids from GenBank are shown in Supplementary Table S2. Sequences of Pet. radiatus from the current study were 99.9%–100% identical with sequences of adult Pet. radiatus from cormorants from Israel (Dzikowski et al., Reference Dzikowski, Levy, Poore, Flowers and Paperna2004) and metacercariae from fish in Hungary (Molnar et al., Reference Molnar, Gibson, Cech, Papp, Deak-Paulus, Juhasz, Toth and Szekely2015). Also, the isolates from the present study showed a close relationship (98.8%–99% similarity) with cercarial isolates from Bi. sudanica in Kenya (Outa et al., Reference Outa, Sattmann, Köhsler, Walochnik and Jirsa2020) and Isi. hainesii from Australia (Barton et al., Reference Barton, Zhu, Nuhoglu, Pearce, McLellan and Shamsi2022). Petasiger sp. 3 ZA differed from Pet. radiatus by p-distances of 5.3%–5.4%. The genetic distance between Petasiger sp. 2 ZA from South African lymnaeid snails (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024) and the current Petasiger spp. ranged between 4.8% and 5.3%. Interestingly, Petasiger sp. 3 ZA had the highest similarity (98.8%–98.9%) with sequences from Tanzania (MZ412883) (Chibwana & Katandukila, Reference Chibwana and Katandukila2021) and Zimbabwe (PP564877) (Mudavanhu et al., Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024) that were published as Stephanoprora amurensis Tatonova, Izrailskaia & Besprozvannykh, 2020 (Echinochasmidae). However, as shown in the supplementary Table S2 and Fig. 13, the two were distant (p-distance = 16%–19%) from other echinochasmid sequences. Therefore, the designation of MZ412883 and PP564877 as S. amurensis (Chibwana & Katandukila, Reference Chibwana and Katandukila2021; Mudavanhu et al., Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024) were erroneous. The p-distances between Echinostomatidae gen. sp. and Petasiger spp. ranged from 4.9% to 8.4% (Supplementary Table S2). ITS rDNA phylogenetic analyses of 39 isolates of echinostomatids (988–1042 bp) revealed that Petasiger spp. grouped in seven strongly supported subclades (Fig. 13). Petasiger radiatus from the present study clustered with three other sequences of Pet. radiatus (subclade A), and the four were sister to subclade B comprising of cercariae of Petasiger from Kenya (and Australia. Petasiger phalacrocoracis sequences formed a single cluster (C) that was basal to D, E, and F. Petasiger sp. 2 ZA from South Africa clustered with sequences of unnamed Petasiger from Hungary and Australia (subclade D). Subclade D was sister to the clade comprising Petasiger sp. 3 ZA and sequences from Tanzania and Zimbabwe which were incorrectly identified as S. amurensis. Sequences of Isthmiophora hortensis (Asada, 1926) and Isthmiophora melis (Schrank, 1788) (subclade G) were nested within the Petasiger clade. Echinostomatidae gen. sp. (subclade J) was monophyletic with sequences of Rhopalias in a moderately supported (0.70%) clade (fig. 13).

Figure 13. Phylogenetic tree based on Bayesian inference (BI) and maximum likelihood (ML) analyses of ITS sequences of Echinostomatidae spp. The clades containing Petasiger and Echinostomatidae gen. sp. are highlighted. Isolates from South Africa are indicated in bold. Nodal support values are given as BI/ML and values lower 0.5 (50%) are not shown. GenBank accession numbers of the sequences are given in parentheses. Isolates MZ412883 (Chibwana & Katandukila, Reference Chibwana and Katandukila2021) and PP564877 (Mudavanhu et al., Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024) marked with asterisks (**), indicate erroneous identification of an unknown Petasiger sp. as Stephanoprora amurensis.
The 18S sequences for each species from the current study were identical. Until now, there were only four 18S rDNA sequences for Petasiger on GenBank, representing Pet. radiatus, Pet. Phalacrocoracis, and an unnamed species. Genetic distances were very low (0%–1.1%) between the present isolates and the representative sequences of Petasiger from GenBank. Consequently, interspecific boundaries were not apparent between some isolates (e.g. Petasiger sp. [Barton et al., Reference Barton, Zhu, Nuhoglu, Pearce, McLellan and Shamsi2022] and Pet. radiatus). In contrast, as shown previously, the two species were clearly distinct in the 28S data. These findings echo previous concerns regarding the unsuitability of using 18S for systematic studies of lower digenean taxonomic groups (Blasco-Costa et al., Reference Blasco-Costa, Cutmore, Miller and Nolan2016). The p-distances between Echinostomatidae gen. sp. and Petasiger spp. ranged from 0.8% to 1.4% (Supplementary Table S3). Phylogenetic reconstruction comprising 23 sequences (876–879 bp) showed that Petasiger isolates were monophyletic. Echinostomatidae gen. sp. was basal to Pegosomum, Isthmiophora and Petasiger (Supplementary Figure S1).
Partial cox1 DNA fragments were generated from three isolates of Petasiger sp. 3 ZA (786–803 bp) and two for Echinostomatidae gen. sp. (632–644 bp). The sequences have been submitted to GenBank as accession numbers PP738976-PP738977 and PP738983-PP738984. Usable sequences were not obtained for Pet. radiatus. Genetic divergence and base pair differences between Petasiger sp. 3 ZA, Echinostomatidae gen. sp. and other echinostomatids, based on cox1 sequences are indicated in Supplementary Table S4. Petasiger sp. 3 ZA haplotypes varied by 0–5 bp, corresponding to p-distances of 0%–1.2%. The p-distances between Petasiger sp. 3 ZA and other Petasiger spp. ranged between 11% and 12%. Cercarial isolates from Zimbabwe that were published as Echinostomata sp. (MT994273-4) and ‘Psilostomidae sp.’ (MT013353) (Schols et al., Reference Schols, Mudavanhu, Carolus, Hammoud, Muzarabani, Barson and Huyse2020), and ‘Stephanoprora amurensis’ (PP556555) (Mudavanhu et al., Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024), showed a close relationship (98.5%–99.8% similarity) with sequences of Petasiger sp. 5 from Uganda (Hammoud et al., Reference Hammoud, Kayenbergh, Tumusiime, Verschuren, Albrecht, Huyse and Van Bocxlaer2022). The small p-distance between the sequences (0.2%–1.5%) suggests that they belong to the same species. Echinostomatidae gen. sp. varied from other echinostomatids by 88–114 bp, corresponding to p-distances of 21.8%–28.3% (Supplementary Table S4). The BI and ML phylograms comprising of 41 echinostomatid sequences (403 bp), demonstrated that Petasiger sp. 3 ZA from South Africa (subclade B) was sister to a cluster (subclade A) comprising the sequences from Zimbabwe and Uganda (Fig. 14). The occurrence of cercarial isolates from Zimbabwe that were designated as ‘Psilostomidae sp.’ and ‘S. amurensis’ (Mudavanhu et al., Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024) within the strongly supported Petasiger clade (Fig. 14), shows that the two cercariae were misidentified. Similarly, sequences from Tanzania designated as Pet. phalacrocoracis (Chibwana & Katandukila, Reference Chibwana and Katandukila2021) clustered with sequences of the family Echinochasmidae (subclades D and E). As mentioned in the ITS results, there appears to be mistakes in the identities of Petasiger and Stephanoprora sequences that were uploaded on GenBank by Chibwana & Katandukila (Reference Chibwana and Katandukila2021). Echinostomatidae gen. sp. formed a branch (subclade C) that was basal to A and B. An unnamed Echinostomata sp. (MT994275) from Physella acuta (Draparnaud, 1805) and Bi. pfeifferi from Zimbabwe showed a close genetic relationship (p-distance = 4.9%) with sequences of Echinostoma miyagawai Ishii, 1932 and they formed a strongly supported clade (F) with other Echinostoma spp. (Fig. 14).

Figure 14. Bayesian inference (BI) and maximum likelihood (ML) phylograms of the relationships between Echinostomatidae spp., based on the cytochrome c oxidase subunit 1 mitochondrial gene (cox1) sequences. The clades containing Petasiger and Echinostomatidae gen. sp. are highlighted. Isolates from South Africa are indicated in bold. The branch length scale indicates the number of substitutions per site. Nodal support values lower than 0.5 (50%) are excluded. Sequences marked with asterisks (**) are for isolates from Zimbabwe (Mudavanhu et al., Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024) that have been synonymised with Petasiger sp. 5 and (***) are from Tanzania (Chibwana & Katandukila, Reference Chibwana and Katandukila2021) whose identities are questionable.
Discussion
In general, morphological descriptions of intramolluscan stages of echinostomatids are often based on cercarial morphological features observed using light microscopy. For Petasiger cercariae, features such as overall body size, number, and arrangement of penetration gland cells and flame cells, number of granules in the main excretory ducts, visibility of the excretory duct in the tail stem, and the presence or absence of tegumental spines are considered when distinguishing between species (Našincová et al., Reference Našincová, Scholz and Moravec1993; King & Van As, Reference King and Van As2000; Fernández et al., Reference Fernández, Hamann and Ostrowski-de Nunez2016; Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). The current study showed that apart from cercariae of Petasiger sp. (Cercaria bruynoghei) and Pet. segregatus, that were distinguishable by their small sized bodies, discrimination between the other species was difficult due to overlap in cercarial dimensions. These findings concur with some studies which showed that cercarial size can be an unreliable criterion for distinguishing between closely related species (Horák et al., Reference Horák, Kolárová and Adema2002; Podhorský et al., Reference Podhorský, Huůzová, Mikeš and Horák2009). Also, differences were observed in body dimensions of Pet. radiatus cercariae from the Czech Republic (Našincová et al., Reference Našincová, Scholz and Moravec1993) and the current study. This variation in cercarial size might have been caused by differences in fixation techniques or it might be indicative of intraspecific variation. Specimens from the present study were fixed in 70% ethanol while the specimens described by Našincová et al. (Reference Našincová, Scholz and Moravec1993) were fixed in 4% formalin. According to Blair & Islam (Reference Blair and Islam1983), fixation can influence the dimensions of cercariae, thereby making it difficult to compare specimens that were fixed using different techniques. In some species, intraspecific variation has been observed in cercariae obtained at different times or from different hosts. For instance, Porter (Reference Porter1938) reported the occurrence of two morphotypes of a sanguinicolid (that differed only in size) from Bul. tropicus collected in different months at the same locality in South Africa. Also, Neuhaus (Reference Neuhaus1952) observed that cercariae of Trichobilharzia szidati Neuhaus, Reference Neuhaus1952 from two lymnaeid species differed significantly in size, despite being collected at the same time and place in Germany and using the same fixation and measurement techniques. Therefore, it is possible that host and environment related factors might have contributed to size differences between the specimens described by Našincová et al. (Reference Našincová, Scholz and Moravec1993) and the current study. Indeed, the current cercariae were isolated from field collected samples of Burnupia spp. (Burnupiidae) while the specimens described by Našincová et al. (Reference Našincová, Scholz and Moravec1993) were obtained from laboratory infected R. auricularia (Lymnaeidae).
In a survey of echinostomes from East Africa, Laidemitt et al. (Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019) used the number of refractile granules in the main excretory ducts to distinguish between cercariae of four Petasiger spp. Based on that criteria, there seems to be three broad groups of cercariae. The first group has very few excretory granules (e.g., Petasiger sp. 2 from Kenya which has 7–10 granules) (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019). The second group has approximately 17–23 granules (e.g., C. decora from DRC, Pet. variospinosus from South Africa, and Petasiger spp. 4 and 5 from Kenya) (Fain, Reference Fain1953; King & Van As, Reference King and Van As2000; Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019). We suggest the inclusion of Petasiger sp. 1 ZA in this second group. Although the number of excretory granules was not mentioned, the photomicrograph provided for Petasiger sp. 1 ZA cercaria showed 20 and 23 granules in the main excretory ducts in the paper by Moema et al. (Reference Moema, King and Baker2008). Based on the presence of 19–20 excretory granules, Laidemitt et al. (Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019) implied that C. decora, Pet. variospinosus, and Petasiger sp. 5 might be identical. In addition, the excretory systems of C. decora, Pet. variospinosus and Petasiger sp. 1 ZA have 28 flame cells (Fain, Reference Fain1953; King & Van As, Reference King and Van As2000; Moema et al., Reference Moema, King and Baker2008). Data for flame cell patterns are not available for Petasiger spp. 4 and 5 (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019). Despite the similarities within this second group, there are some differences that are worth noting. Cercaria decora is distinguished by a cluster of numerous penetration gland cells along the oesophagus, whereas in the other species, penetration gland cells were not reported. Petasiger sp. 1 ZA and Pet. variospinosus are distinguishable based on the number of cilia on their oral papillae. Therefore, synonymity between the cercariae in this second group is unlikely. The third group is composed of Petasiger spp. with numerous granules (>25). For example, Pet. radiatus and Petasiger sp. 3 ZA from the current study, Petasiger sp. 2 ZA (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024), C. bruynoghei (Fain Reference Fain1953), Pet. segregatus (Lie & Basch, Reference Lie and Basch1967), Echinocercaria III (Ostrowski de Núñez et al., Reference de Núñez M, Hamann and Rumi1991) and Petasiger sp. (Fernández et al., Reference Fernández, Hamann and Ostrowski-de Nunez2016). Flame cells were discernible in C. bruynoghei (24) and Echinocercaria III (28), and poorly visible in Petasiger sp. (Fernández et al., Reference Fernández, Hamann and Ostrowski-de Nunez2016), Pet. segregatus, Petasiger sp. 2 ZA, and Petasiger sp. 3 ZA. Further distinctions were based on the visibility and number of penetration gland cells along the oesophagus. Poor visibility of cercarial internal structures usually corresponds to the presence of numerous cystogenous glands (Lie & Basch, Reference Lie and Basch1967; Fernández et al., Reference Fernández, Hamann and Ostrowski-de Nunez2016; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). The close resemblance between Petasiger segregatus from Brazil (Lie & Basch, Reference Lie and Basch1967) and Petasiger sp. from Argentina (Fernández et al., Reference Fernández, Hamann and Ostrowski-de Nunez2016) suggests that they might be identical. Indeed, the number of granules in each excretory duct of Pet. segregatus (40–50) corresponds with Petasiger sp. (45–59). In addition, both species are characterised by sensory hairs (visible using light microscope) and the presence of numerous cystogenous gland cells.
Regarding caudal features, a bifurcated excretory duct near the base of the tail stem is a feature that seems to be limited to two unidentified species from Europe (Kiseliene, Reference Kiseliene1970; Faltýnková et al., Reference Faltýnková, Nasincová and Kablásková2008b). It is also worth noting that Barton et al. (Reference Barton, Zhu, Nuhoglu, Pearce, McLellan and Shamsi2022) mentioned the presence of finfolds along the cercarial tail of Petasiger sp. from Australia. However, several studies have shown that cercariae of Petasiger lack finfolds on their tails (Lie & Basch, Reference Lie and Basch1967; Našincová et al., Reference Našincová, Scholz and Moravec1993; King & Van As, Reference King and Van As2000; Faltýnková et al., Reference Faltýnková, Nasincová and Kablásková2008b; Fernández et al., Reference Fernández, Hamann and Ostrowski-de Nunez2016; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). Indeed, the photomicrograph and drawing provided by Barton et al. (Reference Barton, Zhu, Nuhoglu, Pearce, McLellan and Shamsi2022) only show the lateral sides of the tail trunk which might have been confused for finfolds. Tegumental features of cercariae such as the presence (and density) or absence of sensory hairs, and the patterns of uniciliated and multiciliated papillae, proved to be important for Petasiger species characterisation. However, information of the papillary patterns is available only for Pet. variospinosus (King & Van As, Reference King and Van As2000), Petasiger sp. 1 ZA (Moema et al., Reference Moema, King and Baker2008), Petasiger sp. 2 ZA (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024), and Pet. radiatus and Petasiger sp. 3 ZA (current study). Therefore, there is need to examine more Petasiger species to further demonstrate the usefulness of papillary patterns for species discrimination. Overall, the current study shows that differentiation between species of Petasiger based on cercarial morphology requires the consideration of multiple criteria. Hence, features such as the numbers of refractile granules in the excretory system and the patterns of flame cells, penetration gland cells and papillae, may not be useful for species discrimination when used in isolation.
Apart from the present study, surface features of rediae have been described only for Petasiger sp. 2 ZA (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). Rediae of Petasiger sp. 2 ZA and the current species are characterised by numerous sensilla around the mouth. The presence of oral sensilla seems to be a general feature of most echinostomatids since they have also been reported on Echinostoma paraensei Lie & Basch, Reference Lie and Basch1967 (Pinheiro et al., Reference Pinheiro, Maldonado Junior, Attias and Lanfredi2004) and Ribeiroia ondatrae (Price, 1931) (Keeler et al., Reference Keeler, Fried and Huffman2012). However, the presence and number of multiciliated papillae around the mouth appears to be species specific. For instance, redia of Petasiger sp. 3 ZA was characterised by three pairs of multiciliated papillae while Echinostomatidae gen. sp. had only one pair. In contrast, multiciliated papillae were not reported on the teguments of Ec. paraensei and Ri. ondatrae rediae (Pinheiro et al., Reference Pinheiro, Maldonado Junior, Attias and Lanfredi2004; Keeler et al., Reference Keeler, Fried and Huffman2012).
Molecular data based on 28S rDNA sequences confirmed the placement of the current specimens into the family Echinostomatidae. The identities of cercarial isolates of Pet. radiatus were confirmed based on the 99.9%–100% similarity to sequences of adult worms that were published by Tkach et al. (2016). Petasiger sp. 3 ZA showed a close genetic relationship with cercaria of Petasiger sp. 5 from Kenya (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019). However, the two formed strongly supported divergent lineages with 28S p-distances of 0.6%–0.7%. The divergence was also seen in the cox1 sequences (11.4%–13.2%); hence, corroborating the distinction between Petasiger sp. 3 ZA and Petasiger sp. 5. Cercarial isolate of Petasiger sp. from Australia (OM305105) (Barton et al., Reference Barton, Zhu, Nuhoglu, Pearce, McLellan and Shamsi2022) formed a strongly supported subclade with Petasiger sp. 4 from Bi. sudanica from Lake Victoria, Kenya (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019). Based on this strong genetic relationship, we suggest that the two isolates are haplotypes of the same species. Petasiger sp. metacercariae from Hungary (Cech et al., Reference Cech, Molnár and Székely2017), and cercariae of Petasiger sp. 3 from Kenya (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019) and Petasiger sp. 2 ZA from South Africa (Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024) differed by only (0%–0.1%); hence, they are regarded to represent the same species. The current analyses also confirmed the designation of Petasiger spp. 2 and 6 from Kenya (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019) as distinct species. The high 28S similarity (99.3%–99.5%) between cercaria of Petasiger sp. 1 from Kenya (Laidemitt et al. (Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019) and sequences of Pegosomum asperum and Peg. saginatum (adults) that were obtained from the gall bladder of egret Ardea alba, suggests that these three isolates belong to the same genus. According to Laidemitt et al. (Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019), apart from the number of collar spines (27), other morphological features of Petasiger sp. 1 specimens were obscure since they had been preserved for many years. Similar to Petasiger, Pegosomum spp. are also characterised by 27 collar spines (Heneberg & Sitko, Reference Heneberg and Sitko2017). Since the morphological identification of Petasiger sp. 1 was based only on one cercarial feature, we suspect that the cercaria may have been misidentified.
In the present study, we also incorporated ITS and cox1 sequences of Petasiger (from GenBank), for which 28S data are lacking. For ITS, there is a sequence (MN745952) for cercaria (putatively identified as Pet. variospinosus) that was isolated from Bi. sudanica from Lake Victoria, Kenya (Outa et al., Reference Outa, Sattmann, Köhsler, Walochnik and Jirsa2020). The sequence showed a high similarity (99.6 %) with cercaria of Petasiger sp. from Australia (OM305105) (Barton et al., Reference Barton, Zhu, Nuhoglu, Pearce, McLellan and Shamsi2022) and the two formed a strongly supported subclade. As shown in the 28S data (previous), sequence OM305105 (Barton et al. Reference Barton, Zhu, Nuhoglu, Pearce, McLellan and Shamsi2022) seems to be synonymous with Petasiger sp. 4, also from Bi. sudanica from Lake Victoria, Kenya (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019). Therefore, we suggest that Petasiger cf. variospinosus (Outa et al., Reference Outa, Sattmann, Köhsler, Walochnik and Jirsa2020), Petasiger sp. (Barton et al., Reference Barton, Zhu, Nuhoglu, Pearce, McLellan and Shamsi2022) and Petasiger sp. 4 (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019), belong to the same species. The other ITS sequences (MZ412883 and PP564877) (Chibwana & Katandukila, Reference Chibwana and Katandukila2021; Mudavanhu et al., Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024) are for isolates from Tanzania and Zimbabwe that were labelled as S. amurensis. However, phylogenetic data inferred that MZ412883 and PP564877 belong to Petasiger. As discussed in cox1 data that follows, we suggest that those two sequences that were published by Chibwana & Katandukila (Reference Chibwana and Katandukila2021) and Mudavanhu et al. (Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024) are synonymous with Petasiger sp. 5. Prior to the current study, cox1 sequences (on GenBank) designated as Petasiger spp. were available from two other investigations. The first study published three sequences that were assigned to Pet. phalacrocoracis (Chibwana & Katandukila, Reference Chibwana and Katandukila2021). However, as mentioned in the Results, those sequences clustered with echinochasmids from the same study; hence, their valid identities are uncertain. The second study reported Petasiger sp. from Bul. tropicus in Uganda (Hammoud et al., Reference Hammoud, Kayenbergh, Tumusiime, Verschuren, Albrecht, Huyse and Van Bocxlaer2022). Hammoud et al. (Reference Hammoud, Kayenbergh, Tumusiime, Verschuren, Albrecht, Huyse and Van Bocxlaer2022) noted that the isolates from Uganda were synonymous with Petasiger sp. 5 from Kenya (Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019) based on nad1 sequences. The current study has shown that isolates that were published as Echinostomata sp. (MT994273-4) (Schols et al., Reference Schols, Mudavanhu, Carolus, Hammoud, Muzarabani, Barson and Huyse2020), ‘S. amurensis’ (PP556555) (Mudavanhu et al., Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024) and ‘Psilostomidae sp.’ (MT013353), from Bulinus spp. from Zimbabwe, are haplotypes of Petasiger sp. 5. The sequences of echinostomes that were published by Schols et al. (Reference Schols, Mudavanhu, Carolus, Hammoud, Muzarabani, Barson and Huyse2020) and Mudavanhu et al. (Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024) were from cercariae for which morphological data were not provided. We echo the recommendations of previous studies on the importance of integrated characterisation of cercariae to increase the accuracy of identification and to provide adequate reference data for future studies (Pantoja et al., Reference Pantoja, Faltýnková, O’Dwyer, Jouet, Skírnisson and Kudlai2021; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024). Based on the current findings, it appears that nuclear and mitochondrial DNA sequences of Petasiger on GenBank are representative of Pet. exaeretus, Pet. phalacrocoracis, Pet. radiatus and six unnamed Petasiger spp.
Data on the localities and genotypes of Petasiger indicate a wide geographical distribution of the genus. This concurs with previous studies regarding the cosmopolitan distribution of Petasiger (Faltýnková et al., Reference Faltýnková, Gibson and Kostadinova2008a; Tkach et al., 2016; Barton et al., Reference Barton, Zhu, Nuhoglu, Pearce, McLellan and Shamsi2022). Adults of Petasiger spp. inhabit the intestines of birds belonging to the families Phalacrocoracidae, Anhingidae, Ciconiidae, and Sulidae (Tkach et al., 2016). Although there are only a few reports of adults of Petasiger in Africa, data from the intramolluscan stages show the hidden diversity of Petasiger spp. It appears that the wide distribution of Petasiger is aided not only by the wide distribution of their definitive hosts, but also by their abilities to use diverse first and second intermediate hosts. Indeed, parthenitae and cercariae of Petasiger spp. have been reported from snails of the families Ampullariidae, Bulinidae, Lymnaeidae, and Planorbidae (King and Van As, Reference King and Van As2000; Laidemitt et al., Reference Laidemitt, Brant, Mutuku, Mkoji and Loker2019; Outa et al., Reference Outa, Sattmann, Köhsler, Walochnik and Jirsa2020; Hammoud et al., Reference Hammoud, Kayenbergh, Tumusiime, Verschuren, Albrecht, Huyse and Van Bocxlaer2022; Outa et al., Reference Outa, Bhika and Avenant-Oldewage2024) and Burnupiidae in the current study. Cercariae of Petasiger exit the first intermediate hosts and develop into encysted metacercariae in amphibians and fish, which are the second intermediate hosts (King and van As, Reference King and Van As2000; Kostadinova, Reference Kostadinova, Jones, Bray and Gibson2005; Cech et al., Reference Cech, Molnár and Székely2017).
Phylogenetic analyses demonstrated that Echinostomatidae gen. sp. was distinct from genera whose molecular data are available on GenBank. Its relationships with the other echinostomatids were best inferred using 28S and ITS rDNA, since there are more sequences on GenBank for these markers. That the present species could not be matched with any genus on GenBank confirms that genetic data is still lacking for some echinostomatid genera. According to the keys for the superfamily Echinostomatoidea that were provided by Tkach et al. (2016), the family Echinostomatidae is composed of 38 genera. However, our search through GenBank for sequences of the most widely used markers (28S and ITS) showed that genetic data is available for less than 20 genera. Therefore, in agreement with previous authors (Tkach et al. 2016; Izrailskaia et al., Reference Izrailskaia, Besprozvannykh and Tatonova2021; Pantoja et al., Reference Pantoja, Faltýnková, O’Dwyer, Jouet, Skírnisson and Kudlai2021), we suggest that an expansion of the genetic database of Echinostomatidae is necessary, to enable the validation of species identities and elucidation of suprageneric phylogenetic relationships.
Supplementary material
The supplementary material for this article can be found at http://doi.org/10.1017/S0022149X24000749.
Author contribution
J. O. O. and A. A.-O. conceptualized the study. J. O. O. conducted field sampling, morphological work, molecular and phylogenetic analyses, and wrote original draft of the article. J. O. O. and A. A.-O. reviewed and edited subsequent versions of the manuscript.
Acknowledgements
We are grateful to Beric Gilbert, Kenneth Matea, Lutfiyya Latief, Mpho Maduenyane, and Quinton Dos Santos for assistance during the field studies. We thank the Spectrum Analytical Facility, Faculty of Science at the University of Johannesburg, South Africa, for providing facilities for light and scanning electron microscopy.
Financial support
This study was funded by the University of Johannesburg, South Africa, through the University Research Committee (URC) Postdoctoral Fellowship for JOO, co-funded by the Dean of the Faculty of Science, and AA-O (Research trust fund). Running expenses were funded by the University of Johannesburg, Central Research Committee (URC) and Faculty Research Committee (FRC) to AA-O.
Ethical standard
This research was undertaken following approval by the University of Johannesburg Ethics Committee (Reference Number: 2022-08-05/Outa_Oldewage) and complied with the South African national standard for care and use of animals for scientific purposes.